Epigenetic reprogramming of cancer cells via targeted DNA methylation
- PMID: 22419067
- PMCID: PMC3368819
- DOI: 10.4161/epi.19507
Epigenetic reprogramming of cancer cells via targeted DNA methylation
Abstract
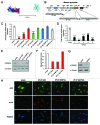
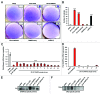
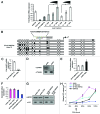
An obstacle in the treatment of human diseases such as cancer is the inability to selectively and effectively target historically undruggable targets such as transcription factors. Here, we employ a novel technology using artificial transcription factors (ATFs) to epigenetically target gene expression in cancer cells. We show that site-specific DNA methylation and long-term stable repression of the tumor suppressor Maspin and the oncogene SOX2 can be achieved in breast cancer cells via zinc-finger ATFs targeting DNA methyltransferase 3a (DNMT3a) to the promoters of these genes. Using this approach, we show Maspin and SOX2 downregulation is more significant as compared with transient knockdown, which is also accompanied by stable phenotypic reprogramming of the cancer cell. These findings indicate that multimodular Zinc Finger Proteins linked to epigenetic editing domains can be used as novel cell resources to selectively and heritably alter gene expression patterns to stably reprogram cell fate.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
