Antibody detection of translocations in Ewing sarcoma
- PMID: 22419563
- PMCID: PMC3443953
- DOI: 10.1002/emmm.201200225
Antibody detection of translocations in Ewing sarcoma
Abstract
The detection of chromosomal translocations has important implications in the diagnosis, prognosis and treatment of patients with cancer. Current approaches to translocation detection have significant shortcomings, including limited sensitivity and/or specificity, and difficulty in application to formalin-fixed paraffin-embedded (FFPE) clinical samples. We developed a new approach called antibody detection of translocations (ADOT) that avoids the shortcomings of current techniques. ADOT combines a transcriptional microarray-based approach with a novel antibody-based detection method. ADOT allows for the accurate and sensitive identification of translocations and provides exon-level information about the fusion transcript. ADOT can detect translocations in poor-quality unprocessed total ribonucleic acid (RNA). Furthermore, the technique is readily generalizable to detect any potential fusion transcript, including previously undescribed fusions. We demonstrate the feasibility of ADOT by examples in which both known and unknown Ewing sarcoma translocations are identified from cell lines, tumour xenografts and FFPE primary tumours. These results demonstrate that ADOT may be an effective approach for translocation analysis in clinical specimens with significant RNA degradation and may offer a novel diagnostic tool for translocation-based cancers.
Copyright © 2012 EMBO Molecular Medicine.
Figures

Ten different EWS/FLI isoforms are represented by fusions between different EWS and FLI exons.
The ADOT approach detects hybrids between RNA transcripts from samples and DNA oligonucleotides on the microarray. These RNA–DNA duplexes are recognized by antibody S9.6 which is then detected by PE-labelled secondary antibody.

ADOT detects translocation transcripts in RNA from 293 cells overexpressing the EWS/FLI 7/6 fusion. The central heatmap shows signal intensity of fusion probes, while the left and bottom heatmaps show those of EWSR1 and FLI exon (Ex) and splice (Spl) probes, respectively. Signal intensities are shown in colour scale. In this case, high signal intensity at row 7, column 6, indicates a fusion between EWSR1 exon 7 and FLI1 exon 6.
Optimization of probe length and signal-to-noise ratio for ADOT. Fusion probes of increasing length as indicated were printed on microarray and hybridized to total RNA from EWS/FLI 7/6 overexpressing 293 cells. Signal-to-noise ratio was then calculated and plotted as a function of probe length.

Heatmaps indicate that A673 cells contain EWS/FLI 7/6 translocation (left); RDES cells harbour EWS/FLI 7/5 translocation (middle); TC466 cells contain EWS/ERG 7/8 translocation (right).
ST97-894 cells were identified by ADOT to contain an EWS/FLI 10/8 translocation.
RT-PCR and sequencing analysis confirming the translocation detected in ST97-894 cells.

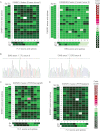
A. Heatmaps showing two frozen tumours that contains an EWS/FLI 7/5 and an EWS/ERG 7/8 translocation, respectively.
B. RT-PCR and sequencing analysis confirming translocation types in frozen tumours tested in A.
C, D. Heatmaps showing EWS/FLI 7/6 translocation in FFPE xenograft (C) and patient tumour (D).
Comment in
-
Molecular investigation of Ewing sarcoma: about detecting translocations.EMBO Mol Med. 2012 Jun;4(6):449-50. doi: 10.1002/emmm.201200226. Epub 2012 Mar 29. EMBO Mol Med. 2012. PMID: 22467249 Free PMC article. No abstract available.
References
-
- Boguslawski SJ, Smith DE, Michalak MA, Mickelson KE, Yehle CO, Patterson WL, Carrico RJ. Characterization of monoclonal antibody to DNA–RNA and its application to immunodetection of hybrids. J Immunol Methods. 1986;89:123–130. - PubMed
-
- Delattre O, Zucman J, Plougastel B, Desmaze C, Melot T, Peter M, Kovar H, Joubert I, de Jong P, Rouleau G, et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359:162–165. - PubMed
-
- Le Deley MC, Delattre O, Schaefer KL, Burchill SA, Koehler G, Hogendoorn PC, Lion T, Poremba C, Marandet J, Ballet S, et al. Impact of EWS-ETS fusion type on disease progression in Ewing's sarcoma/peripheral primitive neuroectodermal tumor: prospective results from the cooperative Euro-E.W.I.N.G. 99 trial. J Clin Oncol. 2010;28:1982–1988. - PubMed
-
- Lessnick SL, Dacwag CS, Golub TR. The Ewing's sarcoma oncoprotein EWS/FLI induces a p53-dependent growth arrest in primary human fibroblasts. Cancer Cell. 2002;1:393–401. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

