An automaton model for the cell cycle
- PMID: 22419973
- PMCID: PMC3262245
- DOI: 10.1098/rsfs.2010.0009
An automaton model for the cell cycle
Abstract
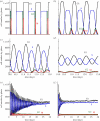
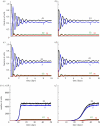
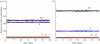
WE CONSIDER AN AUTOMATON MODEL THAT PROGRESSES SPONTANEOUSLY THROUGH THE FOUR SUCCESSIVE PHASES OF THE CELL CYCLE: G1, S (DNA replication), G2 and M (mitosis). Each phase is characterized by a mean duration τ and a variability V. As soon as the prescribed duration of a given phase has passed, the transition to the next phase of the cell cycle occurs. The time at which the transition takes place varies in a random manner according to a distribution of durations of the cell cycle phases. Upon completion of the M phase, the cell divides into two cells, which immediately enter a new cycle in G1. The duration of each phase is reinitialized for the two newborn cells. At each time step in any phase of the cycle, the cell has a certain probability to be marked for exiting the cycle and dying at the nearest G1/S or G2/M transition. To allow for homeostasis, which corresponds to maintenance of the total cell number, we assume that cell death counterbalances cell replication at mitosis. In studying the dynamics of this automaton model, we examine the effect of factors such as the mean durations of the cell cycle phases and their variability, the type of distribution of the durations, the number of cells, the regulation of the cell population size and the independence of steady-state proportions of cells in each phase with respect to initial conditions. We apply the stochastic automaton model for the cell cycle to the progressive desynchronization of cell populations and to their entrainment by the circadian clock. A simple deterministic model leads to the same steady-state proportions of cells in the four phases of the cell cycle.
Keywords: cell cycle; cellular automaton; desynchronization; model; synchronization.
Figures







References
-
- Morgan D. O. 2006. The cell cycle: principles of control. Oxford, UK: Oxford University Press
-
- Goldbeter A. 1991. A minimal cascade model for the mitotic oscillator involving cyclin and cdc2 kinase. Proc. Natl Acad. Sci. USA 88, 9107–911110.1073/pnas.88.20.9107 (doi:10.1073/pnas.88.20.9107) - DOI - DOI - PMC - PubMed
-
- Novak B., Tyson J. J. 1993. Numerical analysis of a comprehensive model of M-phase control in Xenopus oocyte extracts and intact embryos. J. Cell Sci. 106, 1153–1168 - PubMed
-
- Tyson J. J. 1991. Modeling the cell division cycle: cdc2 and cyclin interactions. Proc. Natl Acad. Sci. USA 88, 7328–733210.1073/pnas.88.16.7328 (doi:10.1073/pnas.88.16.7328) - DOI - DOI - PMC - PubMed
-
- Chen K. C., Calzone L., Csikasz-Nagy A., Cross F. R., Novak B., Tyson J. J. 2004. Integrative analysis of cell cycle control in budding yeast. Mol. Biol. Cell 15, 3841–386210.1091/mbc.E03-11-0794 (doi:10.1091/mbc.E03-11-0794) - DOI - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

