Following Ariadne's thread: a new perspective on RBR ubiquitin ligases
- PMID: 22420831
- PMCID: PMC3305615
- DOI: 10.1186/1741-7007-10-24
Following Ariadne's thread: a new perspective on RBR ubiquitin ligases
Abstract
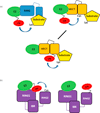
Ubiquitin signaling pathways rely on E3 ligases for effecting the final transfer of ubiquitin from E2 ubiquitin conjugating enzymes to a protein target. Here we re-evaluate the hybrid RING/HECT mechanism used by the E3 family RING-between-RINGs (RBRs) to transfer ubiquitin to substrates. We place RBRs into the context of current knowledge of HECT and RING E3s. Although not as abundant as the other types of E3s (there are only slightly more than a dozen RBR E3s in the human genome), RBRs are conserved in all eukaryotes and play important roles in biology. Re-evaluation of RBR ligases as RING/HECT E3s provokes new questions and challenges the field.
Figures
References
-
- Li W, Bengtson MH, Ulbrich A, Matsuda A, Reddy VA, Orth A, Chanda SK, Batalov S, Joazeiro CA. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle's dynamics and signaling. PLoS One. 2008;3:e1487. doi: 10.1371/journal.pone.0001487. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

