Chromatin state signatures associated with tissue-specific gene expression and enhancer activity in the embryonic limb
- PMID: 22421546
- PMCID: PMC3371702
- DOI: 10.1101/gr.129817.111
Chromatin state signatures associated with tissue-specific gene expression and enhancer activity in the embryonic limb
Abstract
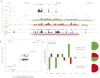
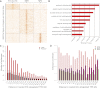
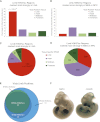
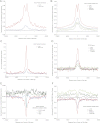
The regulatory elements that direct tissue-specific gene expression in the developing mammalian embryo remain largely unknown. Although chromatin profiling has proven to be a powerful method for mapping regulatory sequences in cultured cells, chromatin states characteristic of active developmental enhancers have not been directly identified in embryonic tissues. Here we use whole-transcriptome analysis coupled with genome-wide profiling of H3K27ac and H3K27me3 to map chromatin states and enhancers in mouse embryonic forelimb and hindlimb. We show that gene-expression differences between forelimb and hindlimb, and between limb and other embryonic cell types, are correlated with tissue-specific H3K27ac signatures at promoters and distal sites. Using H3K27ac profiles, we identified 28,377 putative enhancers, many of which are likely to be limb specific based on strong enrichment near genes highly expressed in the limb and comparisons with tissue-specific EP300 sites and known enhancers. We describe a chromatin state signature associated with active developmental enhancers, defined by high levels of H3K27ac marking, nucleosome displacement, hypersensitivity to sonication, and strong depletion of H3K27me3. We also find that some developmental enhancers exhibit components of this signature, including hypersensitivity, H3K27ac enrichment, and H3K27me3 depletion, at lower levels in tissues in which they are not active. Our results establish histone modification profiling as a tool for developmental enhancer discovery, and suggest that enhancers maintain an open chromatin state in multiple embryonic tissues independent of their activity level.
Figures


References
-
- Bonn S, Zinzen RP, Girardot C, Gustafson EH, Perez-Gonzalez A, Delhomme N, Ghavi-Helm Y, Wilczyński B, Riddell A, Furlong EEM 2012. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat Genet 48: 148–156 - PubMed
-
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441: 349–353 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
