Critical amino acids in Escherichia coli UmuC responsible for sugar discrimination and base-substitution fidelity
- PMID: 22422840
- PMCID: PMC3401427
- DOI: 10.1093/nar/gks233
Critical amino acids in Escherichia coli UmuC responsible for sugar discrimination and base-substitution fidelity
Abstract
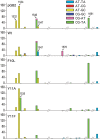
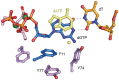
The active form of Escherichia coli DNA polymerase V responsible for damage-induced mutagenesis is a multiprotein complex (UmuD'(2)C-RecA-ATP), called pol V Mut. Optimal activity of pol V Mut in vitro is observed on an SSB-coated single-stranded circular DNA template in the presence of the β/γ complex and a transactivated RecA nucleoprotein filament, RecA*. Remarkably, under these conditions, wild-type pol V Mut efficiently incorporates ribonucleotides into DNA. A Y11A substitution in the 'steric gate' of UmuC further reduces pol V sugar selectivity and converts pol V Mut into a primer-dependent RNA polymerase that is capable of synthesizing long RNAs with a processivity comparable to that of DNA synthesis. Despite such properties, Y11A only promotes low levels of spontaneous mutagenesis in vivo. While the Y11F substitution has a minimal effect on sugar selectivity, it results in an increase in spontaneous mutagenesis. In comparison, an F10L substitution increases sugar selectivity and the overall fidelity of pol V Mut. Molecular modeling analysis reveals that the branched side-chain of L10 impinges on the benzene ring of Y11 so as to constrict its movement and as a consequence, firmly closes the steric gate, which in wild-type enzyme fails to guard against ribonucleoside triphosphates incorporation with sufficient stringency.
Figures







References
-
- Kato T, Shinoura Y. Isolation and characterization of mutants of Escherichia coli deficient in induction of mutations by ultraviolet light. Mol. Gen. Genet. 1977;156:121–131. - PubMed
-
- Steinborn G. Uvm mutants of Escherichia coli K12 deficient in UV mutagenesis. I. Isolation of uvm mutants and their phenotypical characterization in DNA repair and mutagenesis. Mol. Gen. Genet. 1978;165:87–93. - PubMed
-
- Bruck I, Woodgate R, McEntee K, Goodman MF. Purification of a soluble UmuD′C complex from Escherichia coli: cooperative binding of UmuD′C to single-stranded DNA. J. Biol. Chem. 1996;271:10767–10774. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

