Structural basis for the rescue of stalled ribosomes: structure of YaeJ bound to the ribosome
- PMID: 22422986
- PMCID: PMC3377438
- DOI: 10.1126/science.1217443
Structural basis for the rescue of stalled ribosomes: structure of YaeJ bound to the ribosome
Abstract
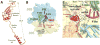
In bacteria, the hybrid transfer-messenger RNA (tmRNA) rescues ribosomes stalled on defective messenger RNAs (mRNAs). However, certain gram-negative bacteria have evolved proteins that are capable of rescuing stalled ribosomes in a tmRNA-independent manner. Here, we report a 3.2 angstrom-resolution crystal structure of the rescue factor YaeJ bound to the Thermus thermophilus 70S ribosome in complex with the initiator tRNA(i)(fMet) and a short mRNA. The structure reveals that the C-terminal tail of YaeJ functions as a sensor to discriminate between stalled and actively translating ribosomes by binding in the mRNA entry channel downstream of the A site between the head and shoulder of the 30S subunit. This allows the N-terminal globular domain to sample different conformations, so that its conserved GGQ motif is optimally positioned to catalyze the hydrolysis of peptidyl-tRNA. This structure gives insights into the mechanism of YaeJ function and provides a basis for understanding how it rescues stalled ribosomes.
Figures
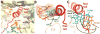

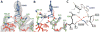
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

