Organismal propagation in the absence of a functional telomerase pathway in Caenorhabditis elegans
- PMID: 22425786
- PMCID: PMC3343340
- DOI: 10.1038/emboj.2012.61
Organismal propagation in the absence of a functional telomerase pathway in Caenorhabditis elegans
Abstract
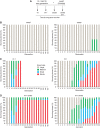
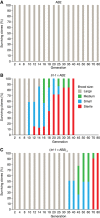
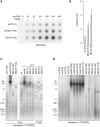
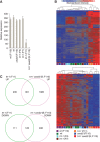
To counteract replication-dependent telomere shortening most eukaryotic cells rely on the telomerase pathway, which is crucial for the maintenance of proliferative potential of germ and stem cell populations of multicellular organisms. Likewise, cancer cells usually engage the telomerase pathway for telomere maintenance to gain immortality. However, in ∼10% of human cancers telomeres are maintained through telomerase-independent alternative lengthening of telomeres (ALT) pathways. Here, we describe the generation and characterization of C. elegans survivors in a strain lacking the catalytic subunit of telomerase and the nematode telomere-binding protein CeOB2. These clonal strains, some of which have been propagated for >180 generations, represent the first example of a multicellular organism with canonical telomeres that can survive without a functional telomerase pathway. The animals display the heterogeneous telomere length characteristic for ALT cells, contain single-stranded C-circles, a transcription profile pointing towards an adaptation to chronic stress and are therefore a unique and valuable tool to decipher the ALT mechanism.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
-
- Blackburn EH (1992) Telomerases. Annu Rev Biochem 61: 113–129 - PubMed
-
- Blasco MA, Lee HW, Hande MP, Samper E, Lansdorp PM, DePinho RA, Greider CW (1997) Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell 91: 25–34 - PubMed
-
- Bryan TM, Englezou A, Dalla-Pozza L, Dunham MA, Reddel RR (1997) Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines [see comments]. Nat Med 3: 1271–1274 - PubMed
-
- de Lange T (1998) Telomeres and senescence: ending the debate. Science 279: 334–335 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

