Network physiology reveals relations between network topology and physiological function
- PMID: 22426223
- PMCID: PMC3518900
- DOI: 10.1038/ncomms1705
Network physiology reveals relations between network topology and physiological function
Abstract
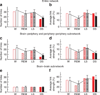
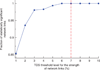
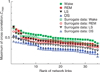
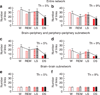
The human organism is an integrated network where complex physiological systems, each with its own regulatory mechanisms, continuously interact, and where failure of one system can trigger a breakdown of the entire network. Identifying and quantifying dynamical networks of diverse systems with different types of interactions is a challenge. Here we develop a framework to probe interactions among diverse systems, and we identify a physiological network. We find that each physiological state is characterized by a specific network structure, demonstrating a robust interplay between network topology and function. Across physiological states, the network undergoes topological transitions associated with fast reorganization of physiological interactions on time scales of a few minutes, indicating high network flexibility in response to perturbations. The proposed system-wide integrative approach may facilitate the development of a new field, Network Physiology.
Figures


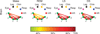
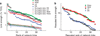

References
-
- Bassingthwaighte JB, Liebovitch L, West BJ. Fractal Physiology. Oxford University Press; 1994.
-
- West GB, Brown JH, Enquist BJ. A general model for the origin of allometric scaling laws in biology. Science. 1997;276:122–126. - PubMed
-
- Bunde A, et al. Correlated and uncorrelated regions in heart-rate fluctuations during sleep. Phys. Rev. Lett. 2000;85:3736–3739. - PubMed
-
- Dvir I, Adler Y, Freimark D, Lavie P. Evidence for fractal correlation properties in variations of peripheral arterial tone during REM sleep. Am. J. Physiol. Heart Circ. Physiol. 2002;283:H434–H439. - PubMed
-
- Ivanov PCh, et al. Scaling behaviour of heartbeat intervals obtained by wavelet-based time-series analysis. Nature. 1996;383:323–327. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

