Diversity and roles of (t)RNA ligases
- PMID: 22426497
- PMCID: PMC3400036
- DOI: 10.1007/s00018-012-0944-2
Diversity and roles of (t)RNA ligases
Abstract
The discovery of discontiguous tRNA genes triggered studies dissecting the process of tRNA splicing. As a result, we have gained detailed mechanistic knowledge on enzymatic removal of tRNA introns catalyzed by endonuclease and ligase proteins. In addition to the elucidation of tRNA processing, these studies facilitated the discovery of additional functions of RNA ligases such as RNA repair and non-conventional mRNA splicing events. Recently, the identification of a new type of RNA ligases in bacteria, archaea, and humans closed a long-standing gap in the field of tRNA processing. This review summarizes past and recent findings in the field of tRNA splicing with a focus on RNA ligation as it preferentially occurs in archaea and humans. In addition to providing an integrated view of the types and phyletic distribution of RNA ligase proteins known to date, this survey also aims at highlighting known and potential accessory biological functions of RNA ligases.
Figures

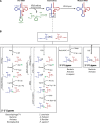


References
-
- Hartmann RK, Gossringer M, Spath B, Fischer S, Marchfelder A. The making of tRNAs and more—RNase P and tRNase Z. Prog Mol Biol Transl Sci. 2009;85:319–368. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

