Identification and characterization of ToRC, a novel ISWI-containing ATP-dependent chromatin assembly complex
- PMID: 22426536
- PMCID: PMC3315121
- DOI: 10.1101/gad.180604.111
Identification and characterization of ToRC, a novel ISWI-containing ATP-dependent chromatin assembly complex
Abstract
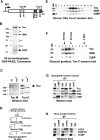
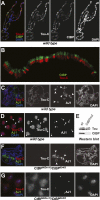
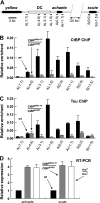
SNF2-like motor proteins, such as ISWI, cooperate with histone chaperones in the assembly and remodeling of chromatin. Here we describe a novel, evolutionarily conserved, ISWI-containing complex termed ToRC (Toutatis-containing chromatin remodeling complex). ToRC comprises ISWI, Toutatis/TIP5 (TTF-I-interacting protein 5), and the transcriptional corepressor CtBP (C-terminal-binding protein). ToRC facilitates ATP-dependent nucleosome assembly in vitro. All three subunits are required for its maximal biochemical activity. The toutatis gene exhibits strong synthetic lethal interactions with CtBP. Thus, ToRC mediates, at least in part, biological activities of CtBP and Toutatis. ToRC subunits colocalize in euchromatic arms of polytene chromosomes. Furthermore, nuclear localization and precise distribution of ToRC in chromosomes are dependent on CtBP. ToRC is involved in CtBP-mediated regulation of transcription by RNA polymerase II in vivo. For instance, both Toutatis and CtBP are required for repression of genes of a proneural gene cluster, achaete-scute complex (AS-C), in Drosophila larvae. Intriguingly, native C-terminally truncated Toutatis isoforms do not associate with CtBP and localize predominantly to the nucleolus. Thus, Toutatis forms two alternative complexes that have differential distribution and can participate in distinct aspects of nuclear DNA metabolism.
Figures
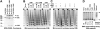
References
-
- Adams CR, Kamakaka RT 1999. Chromatin assembly: Biochemical identities and genetic redundancy. Curr Opin Genet Dev 9: 185–190 - PubMed
-
- Akey CW, Luger K 2003. Histone chaperones and nucleosome assembly. Curr Opin Struct Biol 13: 6–14 - PubMed
-
- Armstrong JA, Bieker JJ, Emerson BM 1998. A SWI/SNF-related chromatin remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro. Cell 95: 93–104 - PubMed
-
- Biryukova I, Heitzler P 2008. Drosophila C-terminal binding protein, dCtBP is required for sensory organ prepattern and sharpens proneural transcriptional activity of the GATA factor Pnr. Dev Biol 323: 64–75 - PubMed
-
- Chinnadurai G 2002. CtBP, an unconventional transcriptional corepressor in development and oncogenesis. Mol Cell 9: 213–224 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
