Establishment of epigenetic patterns in development
- PMID: 22427185
- PMCID: PMC3350763
- DOI: 10.1007/s00412-012-0365-x
Establishment of epigenetic patterns in development
Abstract
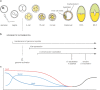
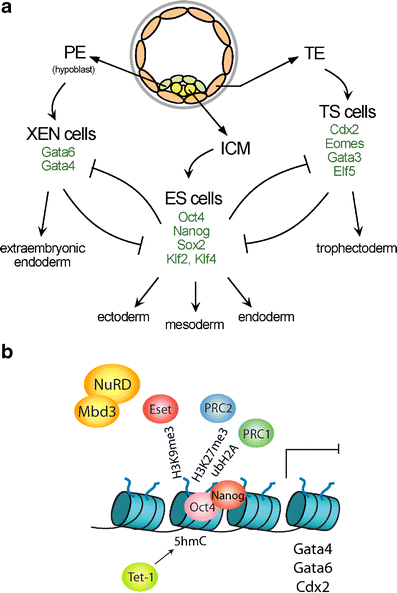
The distinct cell types of the body are established from the fertilized egg in development and assembled into functional tissues. Functional characteristics and gene expression patterns are then faithfully maintained in somatic cell lineages over a lifetime. On the molecular level, transcription factors initiate lineage-specific gene expression programmmes and epigenetic regulation contributes to stabilization of expression patterns. Epigenetic mechanisms are essential for maintaining stable cell identities and their disruption can lead to disease or cellular transformation. Here, we discuss the role of epigenetic regulation in the early mouse embryo, which presents a relatively well-understood system. A number of studies have contributed to the understanding of the function of Polycomb group complexes and the DNA methylation system. The role of many other chromatin regulators in development remains largely unexplored. Albeit the current picture remains incomplete, the view emerges that multiple epigenetic mechanisms cooperate for repressing critical developmental regulators. Some chromatin modifications appear to act in parallel and others might repress the same gene at a different stage of cell differentiation. Studies in pluripotent mouse embryonic stem cells show that epigenetic mechanisms function to repress lineage specific gene expression and prevent extraembryonic differentiation. Insights into this epigenetic "memory" of the first lineage decisions help to provide a better understanding of the function of epigenetic regulation in adult stem cell differentiation.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

