Regulation of Arabidopsis embryo and endosperm development by the polypeptide signaling molecule CLE8
- PMID: 22427333
- PMCID: PMC3336133
- DOI: 10.1105/tpc.111.094839
Regulation of Arabidopsis embryo and endosperm development by the polypeptide signaling molecule CLE8
Abstract
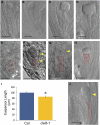
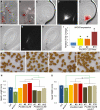
The plant seed is a major nutritional source for humans as well as an essential embryo development and dispersal unit. To ensure proper seed formation, fine spatial and temporal coordination between the embryo, endosperm, and maternal seed components must be achieved. However, the intercellular signaling pathways that direct the synchronous development of these tissues are poorly understood. Here we show that the Arabidopsis thaliana peptide ligand CLAVATA3/embryo surrounding region-related8 (CLE8) is exclusively expressed in young embryos and endosperm, and that it acts cell and noncell autonomously to regulate basal embryo cell division patterns, endosperm proliferation, and the timing of endosperm differentiation. CLE8 positively regulates expression of the transcription factor gene Wuschel-like homeobox8 (WOX8), and together CLE8 and WOX8 form a signaling module that promotes seed growth and overall seed size. These results demonstrate that seed development is coordinated by a secreted peptide ligand that plays a key early role in orchestrating cell patterning and proliferation in the embryo and endosperm.
Figures




References
-
- Bayer M., Nawy T., Giglione C., Galli M., Meinnel T., Lukowitz W. (2009). Paternal control of embryonic patterning in Arabidopsis thaliana. Science 323: 1485–1488 - PubMed
-
- Berger F., Grini P.E., Schnittger A. (2006). Endosperm: An integrator of seed growth and development. Curr. Opin. Plant Biol. 9: 664–670 - PubMed
-
- Brand U., Fletcher J.C., Hobe M., Meyerowitz E.M., Simon R. (2000). Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289: 617–619 - PubMed
-
- Breuninger H., Rikirsch E., Hermann M., Ueda M., Laux T. (2008). Differential expression of WOX genes mediates apical-basal axis formation in the Arabidopsis embryo. Dev. Cell 14: 867–876 - PubMed
-
- Bureau M., Rast M.I., Illmer J., Simon R. (2010). JAGGED LATERAL ORGAN (JLO) controls auxin dependent patterning during development of the Arabidopsis embryo and root. Plant Mol. Biol. 74: 479–491 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

