Leucine-rich repeat and WD repeat-containing protein 1 is recruited to pericentric heterochromatin by trimethylated lysine 9 of histone H3 and maintains heterochromatin silencing
- PMID: 22427655
- PMCID: PMC3340258
- DOI: 10.1074/jbc.M111.337980
Leucine-rich repeat and WD repeat-containing protein 1 is recruited to pericentric heterochromatin by trimethylated lysine 9 of histone H3 and maintains heterochromatin silencing
Abstract
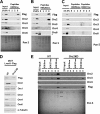
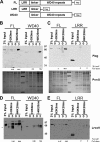
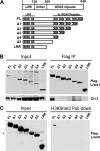
Lrwd1, a protein containing a leucine-rich repeat and a WD40 repeat domain, interacts with the origin replication complex (ORC), a protein complex involved in both initiation of DNA replication and heterochromatin silencing. Lrwd1 and ORC are known to co-purify with repressive histone marks (trimethylated lysine 9 of histone H3 (H3K9me3) and trimethylated lysine 20 of histone H4 (H4K20me3)) and localize to pericentric heterochromatin. However, how the Lrwd1 is recruited to heterochromatin and the functional significance of the localization of Lrwd1 to the heterochromatin are not known. Here, we show that Lrwd1 preferentially binds to trimethylated repressive histone marks in vitro, which is dependent on an intact WD40 domain but independent of ORC proteins. The localization of Lrwd1 and Orc2 at pericentric heterochromatin in mouse cells is lost in cells lacking H3K9me3 but not in cells lacking H4K20me3. In addition, depletion of HP1α has little impact on the localization of Lrwd1 on pericentric heterochromatin. Finally, depletion of Lrwd1 and Orc2 in mouse cells leads to increased transcription of major satellite repeats. These results indicate that the Lrwd1 is recruited to pericentric heterochromatin through binding to H3K9me3 and that the association of Lrwd1 with pericentric heterochromatin is required for heterochromatin silencing and maintenance.
Figures
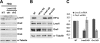


References
-
- Grewal S. I., Jia S. (2007) Heterochromatin revisited. Nat. Rev. Genet. 8, 35–46 - PubMed
-
- Grewal S. I., Moazed D. (2003) Heterochromatin and epigenetic control of gene expression. Science 301, 798–802 - PubMed
-
- Grewal S. I., Elgin S. C. (2002) Heterochromatin: new possibilities for the inheritance of structure. Curr. Opin. Genet. Dev. 12, 178–187 - PubMed
-
- Augui S., Nora E. P., Heard E. (2011) Regulation of X-chromosome inactivation by the X-inactivation centre. Nat. Rev. Genet. 12, 429–442 - PubMed
-
- Wutz A. (2011) Gene silencing in X-chromosome inactivation: advances in understanding facultative heterochromatin formation. Nat. Rev. Genet. 12, 542–553 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

