Quantitative proteomics reveals that only a subset of the endoplasmic reticulum contributes to the phagosome
- PMID: 22427703
- PMCID: PMC3394953
- DOI: 10.1074/mcp.M111.016378
Quantitative proteomics reveals that only a subset of the endoplasmic reticulum contributes to the phagosome
Abstract
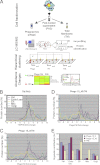
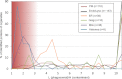
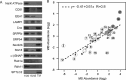
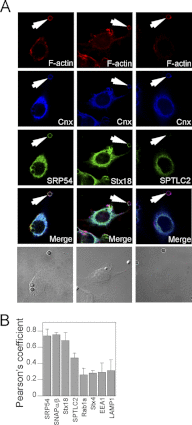
Phagosomes, by killing and degrading pathogens for antigen presentation, are organelles implicated in key aspects of innate and adaptive immunity. Although it has been well established that phagosomes consist of membranes from the plasma membrane, endosomes, and lysosomes, the notion that the endoplasmic reticulum (ER) membrane could play an important role in the formation of the phagosome is debated. However, a method to accurately estimate the contribution of potential source organelles and contaminants to the phagosome proteome has been lacking. Herein, we have developed a proteomic approach for objectively quantifying the contribution of various organelles to the early and late phagosomes by comparing these fractions to their total membrane and postnuclear supernatant of origin in the J774A.1 murine macrophage cell line. Using quantitative label-free mass spectrometry, the abundance of peptides corresponding to hundreds of proteins was estimated and attributed to one of five organelles (e.g. plasma membrane, endosomes/lysosomes, ER, Golgi, and mitochondria). These data in combination with a stable isotope labeling in cell culture method designed to detect potential contaminant sources revealed that the ER is part of the phagosomal membrane and contributes ≈ 20% of the early phagosome proteome. In addition, only a subset of ER proteins is recruited to the phagosome, suggesting that a specific subdomain(s) of the ER might be involved in phagocytosis. Western blotting and immunofluorescence substantially validated this conclusion; we were able to demonstrate that the fraction of the ER in which the ER marker GFP-KDEL accumulates is excluded from the phagosomes, whereas that containing the mVenus-Syntaxin 18 is recruited. These results highlight promising new avenues for the description of the pathogenic mechanisms used by Leishmania, Brucella, and Legionella spp., which thrive in ER-rich phagosomes.
Figures
References
-
- Jutras I., Desjardins M. (2005) Phagocytosis: At the crossroads of innate and adaptive immunity. Annu. Rev. Cell Dev. Biol. 21, 511–527 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

