Metabolic modeling of Chlamydomonas reinhardtii: energy requirements for photoautotrophic growth and maintenance
- PMID: 22427720
- PMCID: PMC3289792
- DOI: 10.1007/s10811-011-9674-3
Metabolic modeling of Chlamydomonas reinhardtii: energy requirements for photoautotrophic growth and maintenance
Abstract
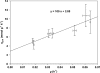
In this study, a metabolic network describing the primary metabolism of Chlamydomonas reinhardtii was constructed. By performing chemostat experiments at different growth rates, energy parameters for maintenance and biomass formation were determined. The chemostats were run at low irradiances resulting in a high biomass yield on light of 1.25 g mol(-1). The ATP requirement for biomass formation from biopolymers (K(x)) was determined to be 109 mmol g(-1) (18.9 mol mol(-1)) and the maintenance requirement (m(ATP)) was determined to be 2.85 mmol g(-1) h(-1). With these energy requirements included in the metabolic network, the network accurately describes the primary metabolism of C. reinhardtii and can be used for modeling of C. reinhardtii growth and metabolism. Simulations confirmed that cultivating microalgae at low growth rates is unfavorable because of the high maintenance requirements which result in low biomass yields. At high light supply rates, biomass yields will decrease due to light saturation effects. Thus, to optimize biomass yield on light energy in photobioreactors, an optimum between low and high light supply rates should be found. These simulations show that metabolic flux analysis can be used as a tool to gain insight into the metabolism of algae and ultimately can be used for the maximization of algal biomass and product yield. ELECTRONIC SUPPLEMENTARY MATERIAL: The online version of this article (doi:10.1007/s10811-011-9674-3) contains supplementary material, which is available to authorized users.
Figures

Similar articles
-
Maximum photosynthetic yield of green microalgae in photobioreactors.Mar Biotechnol (NY). 2010 Nov;12(6):708-18. doi: 10.1007/s10126-010-9258-2. Epub 2010 Feb 23. Mar Biotechnol (NY). 2010. PMID: 20177951 Free PMC article.
-
Effect of O₂:CO₂ ratio on the primary metabolism of Chlamydomonas reinhardtii.Biotechnol Bioeng. 2011 Oct;108(10):2390-402. doi: 10.1002/bit.23194. Epub 2011 May 5. Biotechnol Bioeng. 2011. PMID: 21538341
-
Flux balance analysis of primary metabolism in Chlamydomonas reinhardtii.BMC Syst Biol. 2009 Jan 7;3:4. doi: 10.1186/1752-0509-3-4. BMC Syst Biol. 2009. PMID: 19128495 Free PMC article.
-
Trends on Chlamydomonas reinhardtii growth regimes and bioproducts.Biotechnol Appl Biochem. 2023 Dec;70(6):1830-1842. doi: 10.1002/bab.2486. Epub 2023 Jun 19. Biotechnol Appl Biochem. 2023. PMID: 37337370 Review.
-
Light requirements in microalgal photobioreactors: an overview of biophotonic aspects.Appl Microbiol Biotechnol. 2011 Mar;89(5):1275-88. doi: 10.1007/s00253-010-3047-8. Epub 2010 Dec 23. Appl Microbiol Biotechnol. 2011. PMID: 21181149 Review.
Cited by
-
Reconstruction of the microalga Nannochloropsis salina genome-scale metabolic model with applications to lipid production.BMC Syst Biol. 2017 Jul 4;11(1):66. doi: 10.1186/s12918-017-0441-1. BMC Syst Biol. 2017. PMID: 28676050 Free PMC article.
-
Application of ionizing radiation as an elicitor to enhance the growth and metabolic activities in Chlamydomonas reinhardtii.Front Plant Sci. 2023 Feb 20;14:1087070. doi: 10.3389/fpls.2023.1087070. eCollection 2023. Front Plant Sci. 2023. PMID: 36890890 Free PMC article.
-
Competitive Growth Assay of Mutagenized Chlamydomonas reinhardtii Compatible With the International Space Station Veggie Plant Growth Chamber.Front Plant Sci. 2020 May 25;11:631. doi: 10.3389/fpls.2020.00631. eCollection 2020. Front Plant Sci. 2020. PMID: 32523594 Free PMC article.
-
Impaired Mitochondrial Transcription Termination Disrupts the Stromal Redox Poise in Chlamydomonas.Plant Physiol. 2017 Jul;174(3):1399-1419. doi: 10.1104/pp.16.00946. Epub 2017 May 12. Plant Physiol. 2017. PMID: 28500267 Free PMC article.
-
Metabolic Engineering of Microalgal Based Biofuel Production: Prospects and Challenges.Front Microbiol. 2016 Mar 31;7:432. doi: 10.3389/fmicb.2016.00432. eCollection 2016. Front Microbiol. 2016. PMID: 27065986 Free PMC article. Review.
References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. - PubMed
-
- Ball SG, Dirick L, Decq A, Martiat J-C, Matagne RF. Physiology of starch storage in the monocellular alga Chlamydomonas reinhardtii. Plant Sci. 1990;66(1):1–9. doi: 10.1016/0168-9452(90)90162-H. - DOI
LinkOut - more resources
Full Text Sources
