High-throughput genotoxicity assay identifies antioxidants as inducers of DNA damage response and cell death
- PMID: 22431602
- PMCID: PMC3325711
- DOI: 10.1073/pnas.1114278109
High-throughput genotoxicity assay identifies antioxidants as inducers of DNA damage response and cell death
Abstract
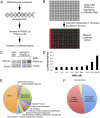
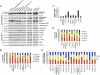
Human ATAD5 is a biomarker for identifying genotoxic compounds because ATAD5 protein levels increase posttranscriptionally in response to DNA damage. We screened over 4,000 compounds with a cell-based quantitative high-throughput ATAD5-luciferase assay detecting genotoxic compounds. We identified 22 antioxidants, including resveratrol, genistein, and baicalein, that are currently used or investigated for the treatment of cardiovascular disease, type 2 diabetes, osteopenia, osteoporosis, and chronic hepatitis, as well as for antiaging. Treatment of dividing cells with these compounds induced DNA damage and resulted in cell death. Despite their genotoxic effects, resveratrol, genistein, and baicalein did not cause mutagenesis, which is a major side effect of conventional anticancer drugs. Furthermore, resveratrol and genistein killed multidrug-resistant cancer cells. We therefore propose that resveratrol, genistein, and baicalein are attractive candidates for improved chemotherapeutic agents.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

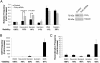
Comment in
-
Oxygen is the key factor associated with the difference between in vivo and in vitro effects of antioxidants.Proc Natl Acad Sci U S A. 2012 Jul 24;109(30):E2028; author reply E2029. doi: 10.1073/pnas.1205916109. Epub 2012 Jun 13. Proc Natl Acad Sci U S A. 2012. PMID: 22699512 Free PMC article. No abstract available.
References
-
- Majka J, Burgers PM. The PCNA-RFC families of DNA clamps and clamp loaders. Prog Nucleic Acid Res Mol Biol. 2004;78:227–260. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

