Composition and similarity of bovine rumen microbiota across individual animals
- PMID: 22432013
- PMCID: PMC3303817
- DOI: 10.1371/journal.pone.0033306
Composition and similarity of bovine rumen microbiota across individual animals
Abstract
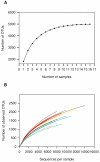
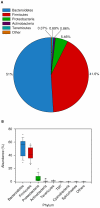
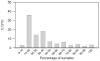
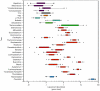
The bovine rumen houses a complex microbiota which is responsible for cattle's remarkable ability to convert indigestible plant mass into food products. Despite this ecosystem's enormous significance for humans, the composition and similarity of bacterial communities across different animals and the possible presence of some bacterial taxa in all animals' rumens have yet to be determined. We characterized the rumen bacterial populations of 16 individual lactating cows using tag amplicon pyrosequencing. Our data showed 51% similarity in bacterial taxa across samples when abundance and occurrence were analyzed using the Bray-Curtis metric. By adding taxon phylogeny to the analysis using a weighted UniFrac metric, the similarity increased to 82%. We also counted 32 genera that are shared by all samples, exhibiting high variability in abundance across samples. Taken together, our results suggest a core microbiome in the bovine rumen. Furthermore, although the bacterial taxa may vary considerably between cow rumens, they appear to be phylogenetically related. This suggests that the functional requirement imposed by the rumen ecological niche selects taxa that potentially share similar genetic features.
Conflict of interest statement
Figures

References
-
- Flint HJ, Bayer EA, Rincon MT, Lamed R, White BA. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat Rev Microbiol. 2008;6:121–131. - PubMed
-
- Flint HJ. The rumen microbial ecosystem–some recent developments. Trends Microbiol. 1997;5:483–488. - PubMed
-
- Welkie DG, Stevenson DM, Weimer PJ. ARISA analysis of ruminal bacterial community dynamics in lactating dairy cows during the feeding cycle. Anaerobe. 2009;16:94–100. - PubMed
-
- Sundset MA, Edwards JE, Cheng YF, Senosiain RS, Fraile MN, et al. Molecular diversity of the rumen microbiome of Norwegian reindeer on natural summer pasture. Microb Ecol. 2009;57:335–348. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

