Functional and molecular characterization of rod-like cells from retinal stem cells derived from the adult ciliary epithelium
- PMID: 22432014
- PMCID: PMC3303820
- DOI: 10.1371/journal.pone.0033338
Functional and molecular characterization of rod-like cells from retinal stem cells derived from the adult ciliary epithelium
Erratum in
- PLoS One. 2012;7(8): doi/10.1371/annotation/ca21f359-8e8e-4c3d-8308-e0f20fc446bb
Abstract
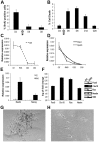
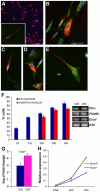
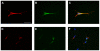
In vitro generation of photoreceptors from stem cells is of great interest for the development of regenerative medicine approaches for patients affected by retinal degeneration and for high throughput drug screens for these diseases. In this study, we show unprecedented high percentages of rod-fated cells from retinal stem cells of the adult ciliary epithelium. Molecular characterization of rod-like cells demonstrates that they lose ciliary epithelial characteristics but acquire photoreceptor features. Rod maturation was evaluated at two levels: gene expression and electrophysiological functionality. Here we present a strong correlation between phototransduction protein expression and functionality of the cells in vitro. We demonstrate that in vitro generated rod-like cells express cGMP-gated channels that are gated by endogenous cGMP. We also identified voltage-gated channels necessary for rod maturation and viability. This level of analysis for the first time provides evidence that adult retinal stem cells can generate highly homogeneous rod-fated cells.
Conflict of interest statement
Figures





Similar articles
-
Cyclic nucleotide-gated ion channels in rod photoreceptors are protected from retinoid inhibition.J Gen Physiol. 2006 Oct;128(4):473-85. doi: 10.1085/jgp.200609619. J Gen Physiol. 2006. PMID: 17001087 Free PMC article.
-
Generation of light-sensitive photoreceptor phenotypes by genetic modification of human adult ocular stem cells with Crx.Invest Ophthalmol Vis Sci. 2010 Feb;51(2):1181-9. doi: 10.1167/iovs.09-4385. Epub 2009 Oct 22. Invest Ophthalmol Vis Sci. 2010. PMID: 19850845
-
Adult ciliary epithelial cells, previously identified as retinal stem cells with potential for retinal repair, fail to differentiate into new rod photoreceptors.Stem Cells. 2010 Jun;28(6):1048-59. doi: 10.1002/stem.423. Stem Cells. 2010. PMID: 20506130
-
Tuning outer segment Ca2+ homeostasis to phototransduction in rods and cones.Adv Exp Med Biol. 2002;514:179-203. doi: 10.1007/978-1-4615-0121-3_11. Adv Exp Med Biol. 2002. PMID: 12596922 Review.
-
cGMP signaling in vertebrate retinal photoreceptor cells.Front Biosci. 2005 May 1;10:1191-204. doi: 10.2741/1612. Front Biosci. 2005. PMID: 15769618 Review.
Cited by
-
Adult Stem Cells, Tools for Repairing the Retina.Curr Ophthalmol Rep. 2019 Mar;7(1):21-29. doi: 10.1007/s40135-019-00195-z. Epub 2019 Jan 24. Curr Ophthalmol Rep. 2019. PMID: 31667009 Free PMC article.
-
Combination of cGMP analogue and drug delivery system provides functional protection in hereditary retinal degeneration.Proc Natl Acad Sci U S A. 2018 Mar 27;115(13):E2997-E3006. doi: 10.1073/pnas.1718792115. Epub 2018 Mar 12. Proc Natl Acad Sci U S A. 2018. PMID: 29531030 Free PMC article.
-
Targeting molecular pathways for the treatment of inherited retinal degeneration.Neural Regen Res. 2020 Oct;15(10):1784-1791. doi: 10.4103/1673-5374.280303. Neural Regen Res. 2020. PMID: 32246618 Free PMC article. Review.
-
A Hyaluronan-Based Injectable Hydrogel Improves the Survival and Integration of Stem Cell Progeny following Transplantation.Stem Cell Reports. 2015 Jun 9;4(6):1031-45. doi: 10.1016/j.stemcr.2015.04.008. Epub 2015 May 14. Stem Cell Reports. 2015. PMID: 25981414 Free PMC article.
-
Photoreceptor cells with profound structural deficits can support useful vision in mice.Invest Ophthalmol Vis Sci. 2014 Mar 25;55(3):1859-66. doi: 10.1167/iovs.13-13661. Invest Ophthalmol Vis Sci. 2014. PMID: 24569582 Free PMC article.
References
-
- Osakada F, Ikeda H, Mandai M, Wataya T, Watanabe K, et al. Toward the generation of rod and cone photoreceptors from mouse, monkey and human embryonic stem cells. Nat Biotechnol. 2008;26:215–224. - PubMed
-
- Zhao X, Liu J, Ahmad I. Differentiation of embryonic stem cells into retinal neurons. Biochem Biophys Res Commun. 2002;297:177–184. - PubMed
-
- Ahmad I, Tang L, Pham H. Identification of neural progenitors in the adult mammalian eye. Biochem Biophys Res Commun. 2000;270:517–521. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

