Integration of GWAS SNPs and tissue specific expression profiling reveal discrete eQTLs for human traits in blood and brain
- PMID: 22433082
- PMCID: PMC3358430
- DOI: 10.1016/j.nbd.2012.03.020
Integration of GWAS SNPs and tissue specific expression profiling reveal discrete eQTLs for human traits in blood and brain
Abstract
Genome-wide association studies have nominated many genetic variants for common human traits, including diseases, but in many cases the underlying biological reason for a trait association is unknown. Subsets of genetic polymorphisms show a statistical association with transcript expression levels, and have therefore been nominated as expression quantitative trait loci (eQTL). However, many tissue and cell types have specific gene expression patterns and so it is not clear how frequently eQTLs found in one tissue type will be replicated in others. In the present study we used two appropriately powered sample series to examine the genetic control of gene expression in blood and brain. We find that while many eQTLs associated with human traits are shared between these two tissues, there are also examples where blood and brain differ, either by restricted gene expression patterns in one tissue or because of differences in how genetic variants are associated with transcript levels. These observations suggest that design of eQTL mapping experiments should consider tissue of interest for the disease or other traits studied.
Published by Elsevier Inc.
Figures
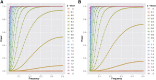
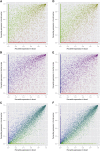

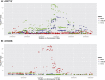
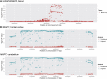
References
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
- G0802462/MRC_/Medical Research Council/United Kingdom
- G-0907/PUK_/Parkinson's UK/United Kingdom
- G0701075/MRC_/Medical Research Council/United Kingdom
- 263 MD 821336/MD/NIMHD NIH HHS/United States
- MR/J006742/1/MRC_/Medical Research Council/United Kingdom
- Z01 AG000185/ImNIH/Intramural NIH HHS/United States
- 263 MD 9164/MD/NIMHD NIH HHS/United States
- G1100616/MRC_/Medical Research Council/United Kingdom
- ZIA AG000947/ImNIH/Intramural NIH HHS/United States
- R01 MD009164/MD/NIMHD NIH HHS/United States
- K01 AG000947/AG/NIA NIH HHS/United States
- G0901254/MRC_/Medical Research Council/United Kingdom
- ZO1-AG000947/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

