Timing and source of subtype-C HIV-1 superinfection in the newly infected partner of Zambian couples with disparate viruses
- PMID: 22433432
- PMCID: PMC3349552
- DOI: 10.1186/1742-4690-9-22
Timing and source of subtype-C HIV-1 superinfection in the newly infected partner of Zambian couples with disparate viruses
Abstract
Background: HIV-1 superinfection occurs at varying frequencies in different at risk populations. Though seroincidence is decreased, in the negative partner of HIV-discordant couples after joint testing and counseling in the Zambia Emory HIV Research Project (ZEHRP) cohort, the annual infection rate remains relatively high at 7-8%. Based on sequencing within the gp41 region of each partner's virus, 24% of new infections between 2004 and 2008 were the result of transmission from a non-spousal partner. Since these seroconvertors and their spouses have disparate epidemiologically-unlinked viruses, there is a risk of superinfection within the marriage. We have, therefore, investigated the incidence and viral origin of superinfection in these couples.
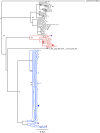
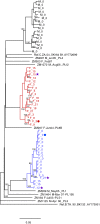
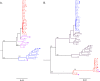
Results: Superinfection was detected by heteroduplex mobility assay (HMA), degenerate base counting of the gp41 sequence, or by phylogenetic analysis of the longitudinal sequences. It was confirmed by full-length env single genome amplification and phylogenetic analysis. In 22 couples (44 individuals), followed for up to five years, three of the newly infected (initially HIV uninfected) partners became superinfected. In each case superinfection occurred during the first 12 months following initial infection of the negative partner, and in each case the superinfecting virus was derived from a non-spousal partner. In addition, one probable case of intra-couple HIV-1 superinfection was observed in a chronically infected partner at the time of his seroconverting spouse's initial viremia. Extensive recombination within the env gene was observed following superinfection.
Conclusions: In this subtype-C discordant couple cohort, superinfection, during the first year after HIV-1 infection of the previously negative partner, occurred at a rate similar to primary infection (13.6% [95% CI 5.2-34.8] vs 7.8% [7.1-8.6]). While limited intra-couple superinfection may in part reflect continued condom usage within couples, this and our lack of detecting newly superinfected individuals after one year of primary infection raise the possibility that immunological resistance to intra-subtype superinfection may develop over time in subtype C infected individuals.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
- AI-51231/AI/NIAID NIH HHS/United States
- KL2 TR000455/TR/NCATS NIH HHS/United States
- MH-66767/MH/NIMH NIH HHS/United States
- HD-40951/HD/NICHD NIH HHS/United States
- T32 AI-007470/AI/NIAID NIH HHS/United States
- R37 AI051231/AI/NIAID NIH HHS/United States
- KL2 RR025009/RR/NCRR NIH HHS/United States
- UL1 TR000454/TR/NCATS NIH HHS/United States
- KL2 RR-025009/RR/NCRR NIH HHS/United States
- P30 AI050409/AI/NIAID NIH HHS/United States
- UL1 RR025008/RR/NCRR NIH HHS/United States
- R01 AI051231/AI/NIAID NIH HHS/United States
- D43 TW001042/TW/FIC NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

