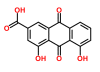
Predicting new molecular targets for rhein using network pharmacology
- PMID: 22433437
- PMCID: PMC3338090
- DOI: 10.1186/1752-0509-6-20
Predicting new molecular targets for rhein using network pharmacology
Retraction in
-
Retraction note: Predicting new molecular targets for rhein using network pharmacology.BMC Syst Biol. 2014;8:105. doi: 10.1186/s12918-014-0105-3. Epub 2014 Sep 18. BMC Syst Biol. 2014. PMID: 25779920 Free PMC article. No abstract available.
Abstract
Background: Drugs can influence the whole biological system by targeting interaction reactions. The existence of interactions between drugs and network reactions suggests a potential way to discover targets. The in silico prediction of potential interactions between drugs and target proteins is of core importance for the identification of new drugs or novel targets for existing drugs. However, only a tiny portion of drug-targets in current datasets are validated interactions. This motivates the need for developing computational methods that predict true interaction pairs with high accuracy. Currently, network pharmacology has used in identifying potential drug targets to predicting the spread of drug activity and greatly contributed toward the analysis of biological systems on a much larger scale than ever before.
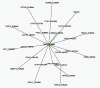
Methods: In this article, we present a computational method to predict targets for rhein by exploring drug-reaction interactions. We have implemented a computational platform that integrates pathway, protein-protein interaction, differentially expressed genome and literature mining data to result in comprehensive networks for drug-target interaction. We used Cytoscape software for prediction rhein-target interactions, to facilitate the drug discovery pipeline.
Results: Results showed that 3 differentially expressed genes confirmed by Cytoscape as the central nodes of the complicated interaction network (99 nodes, 153 edges). Of note, we further observed that the identified targets were found to encompass a variety of biological processes related to immunity, cellular apoptosis, transport, signal transduction, cell growth and proliferation and metabolism.
Conclusions: Our findings demonstrate that network pharmacology can not only speed the wide identification of drug targets but also find new applications for the existing drugs. It also implies the significant contribution of network pharmacology to predict drug targets.
Figures


References
-
- Gilchrist M, Thorsson V, Li B, Rust AG, Korb M, Roach JC, Kennedy K, Hai T, Bolouri H, Aderem A. Systems biology approaches identify ATF3 as a negative regulator of Toll-like receptor. Nature. 2006;441:173-178. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

