Inhibitory mechanism of caspase-6 phosphorylation revealed by crystal structures, molecular dynamics simulations, and biochemical assays
- PMID: 22433863
- PMCID: PMC3346075
- DOI: 10.1074/jbc.M112.351213
Inhibitory mechanism of caspase-6 phosphorylation revealed by crystal structures, molecular dynamics simulations, and biochemical assays
Abstract
The apoptotic effector caspase-6 (CASP6) has been clearly identified as a drug target due to its strong association with neurodegeneration and axonal pruning events as well as its crucial roles in Huntington disease and Alzheimer disease. CASP6 activity is suppressed by ARK5-mediated phosphorylation at Ser(257) with an unclear mechanism. In this work, we solved crystal structures of ΔproCASP6S257E and p20/p10S257E, which mimicked the phosphorylated CASP6 zymogen and activated CASP6, respectively. The structural investigation combined with extensive biochemical assay and molecular dynamics simulation studies revealed that phosphorylation on Ser(257) inhibited self-activation of CASP6 zymogen by "locking" the enzyme in the TEVD(193)-bound "inhibited state." The structural and biochemical results also showed that phosphorylation on Ser(257) inhibited the CASP6 activity by steric hindrance. These results disclosed the inhibition mechanism of CASP6 phosphorylation and laid the foundation for a new strategy of rational CASP6 drug design.
Figures

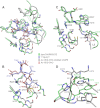


Similar articles
-
Rare human Caspase-6-R65W and Caspase-6-G66R variants identify a novel regulatory region of Caspase-6 activity.Sci Rep. 2018 Mar 13;8(1):4428. doi: 10.1038/s41598-018-22283-z. Sci Rep. 2018. PMID: 29535332 Free PMC article.
-
The regulatory mechanism of the caspase 6 pro-domain revealed by crystal structure and biochemical assays.Acta Crystallogr D Biol Crystallogr. 2014 Jan;70(Pt 1):58-67. doi: 10.1107/S1399004713024218. Epub 2013 Dec 24. Acta Crystallogr D Biol Crystallogr. 2014. PMID: 24419379
-
Crystal structures of human caspase 6 reveal a new mechanism for intramolecular cleavage self-activation.EMBO Rep. 2010 Nov;11(11):841-7. doi: 10.1038/embor.2010.141. Epub 2010 Oct 1. EMBO Rep. 2010. PMID: 20890311 Free PMC article.
-
Caspase-6 and neurodegeneration.Trends Neurosci. 2011 Dec;34(12):646-56. doi: 10.1016/j.tins.2011.09.001. Epub 2011 Oct 22. Trends Neurosci. 2011. PMID: 22018804 Review.
-
Activation and regulation of caspase-6 and its role in neurodegenerative diseases.Annu Rev Pharmacol Toxicol. 2015;55:553-72. doi: 10.1146/annurev-pharmtox-010814-124414. Epub 2014 Oct 17. Annu Rev Pharmacol Toxicol. 2015. PMID: 25340928 Review.
Cited by
-
Tunable allosteric library of caspase-3 identifies coupling between conserved water molecules and conformational selection.Proc Natl Acad Sci U S A. 2016 Oct 11;113(41):E6080-E6088. doi: 10.1073/pnas.1603549113. Epub 2016 Sep 28. Proc Natl Acad Sci U S A. 2016. PMID: 27681633 Free PMC article.
-
Non-Canonical Roles of Apoptotic Caspases in the Nervous System.Front Cell Dev Biol. 2022 Feb 23;10:840023. doi: 10.3389/fcell.2022.840023. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35281082 Free PMC article. Review.
-
Rare human Caspase-6-R65W and Caspase-6-G66R variants identify a novel regulatory region of Caspase-6 activity.Sci Rep. 2018 Mar 13;8(1):4428. doi: 10.1038/s41598-018-22283-z. Sci Rep. 2018. PMID: 29535332 Free PMC article.
-
Structural snapshots reveal distinct mechanisms of procaspase-3 and -7 activation.Proc Natl Acad Sci U S A. 2013 May 21;110(21):8477-82. doi: 10.1073/pnas.1306759110. Epub 2013 May 6. Proc Natl Acad Sci U S A. 2013. PMID: 23650375 Free PMC article.
-
Modifying caspase-3 activity by altering allosteric networks.Biochemistry. 2014 Dec 9;53(48):7582-95. doi: 10.1021/bi500874k. Epub 2014 Nov 21. Biochemistry. 2014. PMID: 25343534 Free PMC article.
References
-
- Yan N., Shi Y. (2005) Mechanisms of apoptosis through structural biology. Annu. Rev. Cell Dev. Biol. 21, 35–56 - PubMed
-
- Slee E. A., Harte M. T., Kluck R. M., Wolf B. B., Casiano C. A., Newmeyer D. D., Wang H. G., Reed J. C., Nicholson D. W., Alnemri E. S., Green D. R., Martin S. J. (1999) Ordering the cytochrome c-initiated caspase cascade: hierarchical activation of caspases-2, -3, -6, -7, -8, and -10 in a caspase-9-dependent manner. J. Cell Biol. 144, 281–292 - PMC - PubMed
-
- Klaiman G., Champagne N., LeBlanc A. C. (2009) Self-activation of caspase-6 in vitro and in vivo: caspase-6 activation does not induce cell death in HEK293T cells. Biochim. Biophys. Acta 1793, 592–601 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

