Interactions of nucleolin and ribosomal protein L26 (RPL26) in translational control of human p53 mRNA
- PMID: 22433872
- PMCID: PMC3351294
- DOI: 10.1074/jbc.M112.349274
Interactions of nucleolin and ribosomal protein L26 (RPL26) in translational control of human p53 mRNA
Abstract
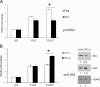
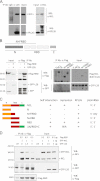
Ribosomal protein RPL26 enhances p53 translation after DNA damage, and this regulation depends upon interactions between the 5'- and 3'-UTRs of human p53 mRNA (Takagi, M., Absalon, M. J., McLure, K. G., and Kastan, M. B. (2005) Cell 123, 49-63; Chen, J., and Kastan, M. B. (2010) Genes Dev. 24, 2146-2156). In contrast, nucleolin (NCL) suppresses the translation of p53 mRNA and its induction after DNA damage. We confirmed reports that RPL26 and NCL interact with each other and then explored the potential role of this interaction in the translational control of p53 after stress. NCL repression of p53 translation utilizes both the 5'- and 3'-UTRs of p53 mRNA, and NCL binds to the same 5'-3'-UTR interaction region that is critical for the recruitment of RPL26 to p53 mRNA after DNA damage. We also found that NCL is able to oligomerize, consistent with a model in which NCL stabilizes this double-stranded RNA structure. We found that the RNA-binding domain of NCL participates in binding to p53 mRNA, is required for both NCL dimerization and NCL-mediated translational repression, and is the domain of NCL that interacts with RPL26. Excessive RPL26 disrupts NCL dimerization, and point mutations in the NCL-interacting region of RPL26 reduce NCL-RPL26 interactions and attenuate both RPL26 binding to human p53 mRNA and p53 induction by RPL26. These observations suggest a model in which the base pairings in the p53 UTR interaction regions are critical for both translational repression and stress induction of p53 by NCL and RPL26, respectively, and that disruption of a NCL-NCL homodimer by RPL26 may be the switch between translational repression and activation after stress.
Figures



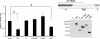

References
-
- Takagi M., Absalon M. J., McLure K. G., Kastan M. B. (2005) Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell 123, 49–63 - PubMed
-
- Kastan M. B., Bartek J. (2004) Cell cycle checkpoints and cancer. Nature 432, 316–323 - PubMed
-
- Ashcroft M., Vousden K. H. (1999) Regulation of p53 stability. Oncogene 18, 7637–7643 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

