The protein gp74 from the bacteriophage HK97 functions as a HNH endonuclease
- PMID: 22434504
- PMCID: PMC3403416
- DOI: 10.1002/pro.2064
The protein gp74 from the bacteriophage HK97 functions as a HNH endonuclease
Abstract
The last gene in the genome of the bacteriophage HK97 encodes the protein gp74. We present data in this article that demonstrates, for the first time, that gp74 possesses HNH endonuclease activity. HNH endonucleases are small DNA binding and digestion proteins characterized by two His residues and an Asn residue. We demonstrate that gp74 cleaves lambda phage DNA at multiple sites and that gp74 requires divalent metals for its endonuclease activity. We also present intrinsic tryptophan fluorescence data that show direct binding of Ni(2+) to gp74. The activity of gp74 in the presence of Ni(2+) is significantly decreased below neutral pH, suggesting the presence of one or more His residues in metal binding and/or DNA digestion. Surprisingly, this pH-dependence of activity is not seen with Zn(2+) , suggesting a different mode of binding of Zn(2+) and Ni(2+) . This difference in activity may result from binding of a second Zn(2+) ion by a putative zinc finger in gp74 in addition to binding of a Zn(2+) ion by the HNH motif. These studies define the biochemical function of gp74 as an HNH endonuclease and provide a platform for determining the role of gp74 in life cycle of the bacteriophage HK97.
Copyright © 2012 The Protein Society.
Figures
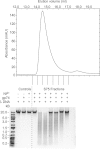


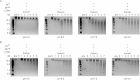
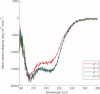
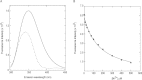
References
-
- Juhala RJ, Ford ME, Duda RL, Youlton A, Hatfull GF, Hendrix RW. Genomic sequences of bacteriophages HK97 and HK022: pervasive genetic mosaicism in the lambdoid bacteriophages. J Mol Biol. 2000;299:27–51. - PubMed
-
- Cardarelli L, Lam R, Tuite A, Baker LA, Sadowski PD, Radford DR, Rubinstein JL, Battaile KP, Chirgadze N, Maxwell KL, Davidson AR. The crystal structure of bacteriophage HK97 gp6: defining a large family of head-tail connector proteins. J Mol Biol. 2010;395:754–768. - PubMed
-
- Mahdi AA, Sharples GJ, Mandal TN, Lloyd RG. Holliday junction resolvases encoded by homologous rusA genes in Escherichia coli K-12 and phage 82. J Mol Biol. 1996;257:561–573. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

