Disease model curation improvements at Mouse Genome Informatics
- PMID: 22434831
- PMCID: PMC3308153
- DOI: 10.1093/database/bar063
Disease model curation improvements at Mouse Genome Informatics
Abstract
Optimal curation of human diseases requires an ontology or structured vocabulary that contains terms familiar to end users, is robust enough to support multiple levels of annotation granularity, is limited to disease terms and is stable enough to avoid extensive reannotation following updates. At Mouse Genome Informatics (MGI), we currently use disease terms from Online Mendelian Inheritance in Man (OMIM) to curate mouse models of human disease. While OMIM provides highly detailed disease records that are familiar to many in the medical community, it lacks structure to support multilevel annotation. To improve disease annotation at MGI, we evaluated the merged Medical Subject Headings (MeSH) and OMIM disease vocabulary created by the Comparative Toxicogenomics Database (CTD) project. Overlaying MeSH onto OMIM provides hierarchical access to broad disease terms, a feature missing from the OMIM. We created an extended version of the vocabulary to meet the genetic disease-specific curation needs at MGI. Here we describe our evaluation of the CTD application, the extensions made by MGI and discuss the strengths and weaknesses of this approach. DATABASE URL: http://www.informatics.jax.org/
Figures
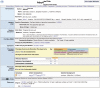


References
-
- Bodenreider O, Burgun A. Towards desiderata for an ontology of diseases for the annotation of biological datasets. ICBO. 2009:39–42.
-
- Iwamoto T, Okumura S, Iwatsubo K, et al. Motor dysfunction in type 5 adenylyl cyclase-null mice. J. Biol. Chem. 2003;278:16936–16940. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

