Grinder: a versatile amplicon and shotgun sequence simulator
- PMID: 22434876
- PMCID: PMC3384353
- DOI: 10.1093/nar/gks251
Grinder: a versatile amplicon and shotgun sequence simulator
Abstract
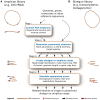
We introduce Grinder (http://sourceforge.net/projects/biogrinder/), an open-source bioinformatic tool to simulate amplicon and shotgun (genomic, metagenomic, transcriptomic and metatranscriptomic) datasets from reference sequences. This is the first tool to simulate amplicon datasets (e.g. 16S rRNA) widely used by microbial ecologists. Grinder can create sequence libraries with a specific community structure, α and β diversities and experimental biases (e.g. chimeras, gene copy number variation) for commonly used sequencing platforms. This versatility allows the creation of simple to complex read datasets necessary for hypothesis testing when developing bioinformatic software, benchmarking existing tools or designing sequence-based experiments. Grinder is particularly useful for simulating clinical or environmental microbial communities and complements the use of in vitro mock communities.
Figures
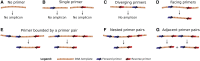

References
-
- Quince C, Lanzén A, Curtis TP, Davenport RJ, Hall N, Head IM, Read LF, Sloan WT. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat. Methods. 2009;6:639–641. - PubMed

