Identification and classification of bacterial Type III toxin-antitoxin systems encoded in chromosomal and plasmid genomes
- PMID: 22434880
- PMCID: PMC3401426
- DOI: 10.1093/nar/gks231
Identification and classification of bacterial Type III toxin-antitoxin systems encoded in chromosomal and plasmid genomes
Abstract
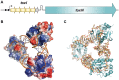
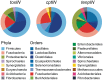
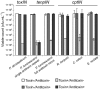
Toxin-antitoxin systems are widespread in bacteria and archaea. They perform diverse functional roles, including the generation of persistence, maintenance of genetic loci and resistance to bacteriophages through abortive infection. Toxin-antitoxin systems have been divided into three types, depending on the nature of the interacting macromolecules. The recently discovered Type III toxin-antitoxin systems encode protein toxins that are inhibited by pseudoknots of antitoxic RNA, encoded by short tandem repeats upstream of the toxin gene. Recent studies have identified the range of Type I and Type II systems within current sequence databases. Here, structure-based homology searches were combined with iterative protein sequence comparisons to obtain a current picture of the prevalence of Type III systems. Three independent Type III families were identified, according to toxin sequence similarity. The three families were found to be far more abundant and widespread than previously known, with examples throughout the Firmicutes, Fusobacteria and Proteobacteria. Functional assays confirmed that representatives from all three families act as toxin-antitoxin loci within Escherichia coli and at least two of the families confer resistance to bacteriophages. This study shows that active Type III toxin-antitoxin systems are far more diverse than previously known, and suggests that more remain to be identified.
Figures


References
-
- Hayes F. Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science. 2003;301:1496–1499. - PubMed
-
- Gerdes K, Christensen SK, Lobner-Olesen A. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 2005;3:371–382. - PubMed
-
- Hayes F, Van Melderen L. Toxins-antitoxins: diversity, evolution and function. Crit. Rev. Biochem. Mol. Biol. 2011;46:386–408. - PubMed
-
- Blower TR, Salmond GP, Luisi BF. Balancing at survival’s edge: the structure and adaptive benefits of prokaryotic toxin-antitoxin partners. Curr. Opin. Struct. Biol. 2011;21:109–118. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

