Phase variable genes of Campylobacter jejuni exhibit high mutation rates and specific mutational patterns but mutability is not the major determinant of population structure during host colonization
- PMID: 22434884
- PMCID: PMC3401435
- DOI: 10.1093/nar/gks246
Phase variable genes of Campylobacter jejuni exhibit high mutation rates and specific mutational patterns but mutability is not the major determinant of population structure during host colonization
Abstract
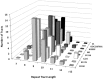
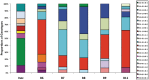
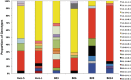
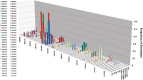
Phase variation of surface structures occurs in diverse bacterial species due to stochastic, high frequency, reversible mutations. Multiple genes of Campylobacter jejuni are subject to phase variable gene expression due to mutations in polyC/G tracts. A modal length of nine repeats was detected for polyC/G tracts within C. jejuni genomes. Switching rates for these tracts were measured using chromosomally-located reporter constructs and high rates were observed for cj1139 (G8) and cj0031 (G9). Alteration of the cj1139 tract from G8 to G11 increased mutability 10-fold and changed the mutational pattern from predominantly insertions to mainly deletions. Using a multiplex PCR, major changes were detected in 'on/off' status for some phase variable genes during passage of C. jejuni in chickens. Utilization of observed switching rates in a stochastic, theoretical model of phase variation demonstrated links between mutability and genetic diversity but could not replicate observed population diversity. We propose that modal repeat numbers have evolved in C. jejuni genomes due to molecular drivers associated with the mutational patterns of these polyC/G repeats, rather than by selection for particular switching rates, and that factors other than mutational drift are responsible for generating genetic diversity during host colonization by this bacterial pathogen.
Figures


References
-
- Bayliss CD. Determinants of phase variation rate and the fitness implications of differing rates for bacterial pathogens and commensals. FEMS Microbiol. Rev. 2009;33:504–520. - PubMed
-
- Moxon ER, Bayliss CD, Hood DW. Bacterial contingency loci: the role of simple sequence DNA repeats in bacterial adaptation. Ann. Rev. Genet. 2007;40:307–333. - PubMed
-
- De Bolle X, Bayliss CD, Field D, van de Ven T, Saunders NJ, Hood DW, Moxon ER. The length of a tetranucleotide repeat tract in Haemophilus influenzae determines the phase variation rate of a gene with homology to type III DNA methyltransferases. Mol. Microbiol. 2000;35:211–222. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

