Comparative genomics of Ceriporiopsis subvermispora and Phanerochaete chrysosporium provide insight into selective ligninolysis
- PMID: 22434909
- PMCID: PMC3325662
- DOI: 10.1073/pnas.1119912109
Comparative genomics of Ceriporiopsis subvermispora and Phanerochaete chrysosporium provide insight into selective ligninolysis
Erratum in
- Proc Natl Acad Sci U S A. 2012 May 22;109(21):8352. San, Ryu Jae [corrected to Ryu, Jae San]
Abstract
Efficient lignin depolymerization is unique to the wood decay basidiomycetes, collectively referred to as white rot fungi. Phanerochaete chrysosporium simultaneously degrades lignin and cellulose, whereas the closely related species, Ceriporiopsis subvermispora, also depolymerizes lignin but may do so with relatively little cellulose degradation. To investigate the basis for selective ligninolysis, we conducted comparative genome analysis of C. subvermispora and P. chrysosporium. Genes encoding manganese peroxidase numbered 13 and five in C. subvermispora and P. chrysosporium, respectively. In addition, the C. subvermispora genome contains at least seven genes predicted to encode laccases, whereas the P. chrysosporium genome contains none. We also observed expansion of the number of C. subvermispora desaturase-encoding genes putatively involved in lipid metabolism. Microarray-based transcriptome analysis showed substantial up-regulation of several desaturase and MnP genes in wood-containing medium. MS identified MnP proteins in C. subvermispora culture filtrates, but none in P. chrysosporium cultures. These results support the importance of MnP and a lignin degradation mechanism whereby cleavage of the dominant nonphenolic structures is mediated by lipid peroxidation products. Two C. subvermispora genes were predicted to encode peroxidases structurally similar to P. chrysosporium lignin peroxidase and, following heterologous expression in Escherichia coli, the enzymes were shown to oxidize high redox potential substrates, but not Mn(2+). Apart from oxidative lignin degradation, we also examined cellulolytic and hemicellulolytic systems in both fungi. In summary, the C. subvermispora genetic inventory and expression patterns exhibit increased oxidoreductase potential and diminished cellulolytic capability relative to P. chrysosporium.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
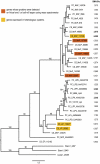

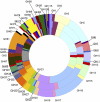
References
-
- United States Department of Energy . Breaking the Biological Barriers to Cellulosic Ethanol: A Joint Research Agenda. Report from the December 2005 Workshop, DOE/SC-0095. Washington, DC: US Department of Energy Office of Science; 2006.
-
- Blanchette R, Krueger E, Haight J, Akhtar M, Akin D. Cell wall alterations in loblolly pine wood decayed by the white-rot fungus, Ceriporiopsis subvermispora. J Biotechnol. 1997;53:203–213.
-
- Hammel KE, Cullen D. Role of fungal peroxidases in biological ligninolysis. Curr Opin Plant Biol. 2008;11:349–355. - PubMed
-
- Martinez AT. Molecular biology and structure-function of lignin-degrading heme peroxidases. Enzyme Microb Technol. 2002;30:425–444.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

