The atypical PKCs in inflammation: NF-κB and beyond
- PMID: 22435553
- PMCID: PMC3531713
- DOI: 10.1111/j.1600-065X.2012.01093.x
The atypical PKCs in inflammation: NF-κB and beyond
Abstract
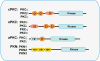
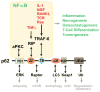
From the very early days of nuclear factor-κB (NF-κB) research, it was recognized that different protein kinase C (PKC) isoforms might be involved in the activation of NF-κB. Pharmacological tools and pseudosubstrate inhibitors suggested that these kinases play a role in this important inflammatory and survival pathway; however, it was the analysis of several genetic mouse knockout models that revealed the complexity and interrelations between the different components of the PB1 network in several cellular functions, including T-cell biology, bone homeostasis, inflammation associated with the metabolic syndrome, and cancer. These studies unveiled, for example, the critical role of PKCζ as a positive regulator of NF-κB through the regulation of RelA but also its inflammatory suppressor activities through the regulation of the interleukin-4 signaling cascade. This observation is of relevance in T cells, where p62, PKCζ, PKCλ/ι, and NBR1 establish a mesh of interactions that culminate in the regulation of T-cell effector responses through the modulation of T-cell polarity. Many questions remain to be answered, not just from the point of view of the implication for NF-κB activation but also with regard to the in vivo interplay between these pathways in pathophysiological processes like obesity and cancer.
© 2012 John Wiley & Sons A/S.
Conflict of interest statement
The authors have no conflicts of interest to declare.
Figures


References
-
- Hanks SK, Hunter T. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 1995;9:576–596. - PubMed
-
- Nishizuka Y. The role of protein kinase C in cell surface signal transduction and tumour promotion. Nature. 1984;308:693–698. - PubMed
-
- Nakamura S, Yamamura H. Yasutomi Nishizuka: father of protein kinase C. J Biochem. 2010;148:125–130. - PubMed
-
- Kikkawa U, Takai Y, Tanaka Y, Miyake R, Nishizuka Y. Protein kinase C as a possible receptor protein of tumor-promoting phorbol esters. J Biol Chem. 1983;258:11442–11445. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

