Chemistry and biology of multicomponent reactions
- PMID: 22435608
- PMCID: PMC3712876
- DOI: 10.1021/cr100233r
Chemistry and biology of multicomponent reactions
Figures


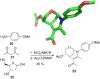
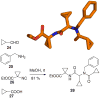
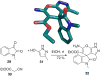
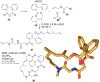
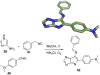
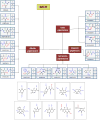
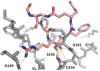
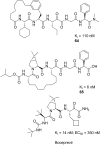


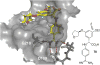

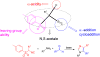
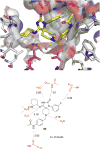
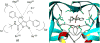
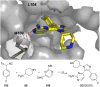
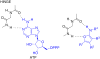

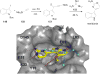


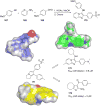
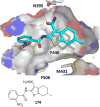
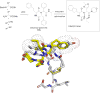
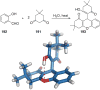
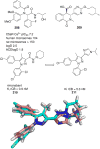
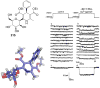



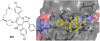
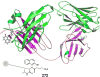
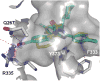
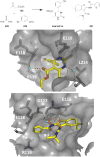
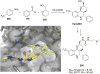
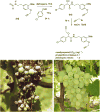
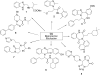
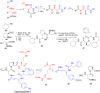

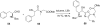

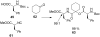


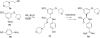



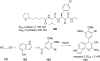
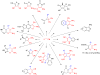
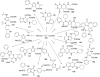
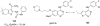

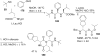
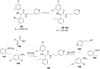
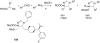
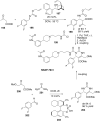

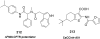
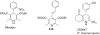

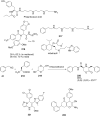
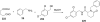
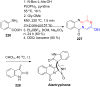
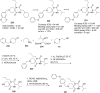

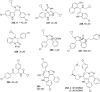
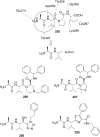
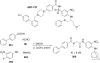
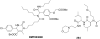
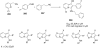
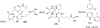
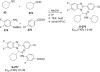
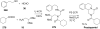


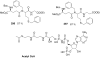
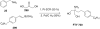
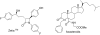
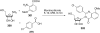


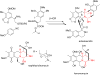
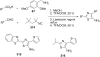
References
-
- Ugi I, Dömling A, Horl W. Endeavour. 1994;18:115.
-
- Tietze LF. Chem Rev. 1996;96:115. - PubMed
-
- Ugi I, Meyr R, Fetzer U, Steinbrückner C. Angew Chem. 1959;71:386.
-
- Passerini M. Gazz Chim Ital. 1921;51:126.
-
- van Leusen D, van Leusen AM. Org React (NY) 2003;57:419.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

