The toxin/immunity network of Burkholderia pseudomallei contact-dependent growth inhibition (CDI) systems
- PMID: 22435733
- PMCID: PMC3331888
- DOI: 10.1111/j.1365-2958.2012.08039.x
The toxin/immunity network of Burkholderia pseudomallei contact-dependent growth inhibition (CDI) systems
Abstract
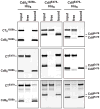
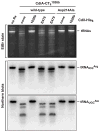
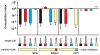
Burkholderia pseudomallei is a category B pathogen and the causative agent of melioidosis--a serious infectious disease that is typically acquired directly from environmental reservoirs. Nearly all B. pseudomallei strains sequenced to date (> 85 isolates) contain gene clusters that are related to the contact-dependent growth inhibition (CDI) systems of γ-proteobacteria. CDI systems from Escherichia coli and Dickeya dadantii play significant roles in bacterial competition, suggesting these systems may also contribute to the competitive fitness of B. pseudomallei. Here, we identify 10 distinct CDI systems in B. pseudomallei based on polymorphisms within the cdiA-CT/cdiI coding regions, which are predicted to encode CdiA-CT/CdiI toxin/immunity protein pairs. Biochemical analysis of three B. pseudomallei CdiA-CTs revealed that each protein possesses a distinct tRNase activity capable of inhibiting cell growth. These toxin activities are blocked by cognate CdiI immunity proteins, which specifically bind the CdiA-CT and protect cells from growth inhibition. Using Burkholderia thailandensis E264 as a model, we show that a CDI system from B. pseudomallei 1026b mediates CDI and is capable of delivering CdiA-CT toxins derived from other B. pseudomallei strains. These results demonstrate that Burkholderia species contain functional CDI systems, which may confer a competitive advantage to these bacteria.
© 2012 Blackwell Publishing Ltd.
Figures






References
-
- Aiyar A, Xiang Y, Leis J. Site-directed mutagenesis using overlap extension PCR. Methods Mol Biol. 1996;57:177–191. - PubMed
-
- Aoki SK, Pamma R, Hernday AD, Bickham JE, Braaten BA, Low DA. Contact-dependent inhibition of growth in Escherichia coli. Science. 2005;309:1245–1248. - PubMed
-
- Beckwith JR, Signer ER. Transposition of the lac region of Escherichia coli. I. Inversion of the lac operon and transduction of lac by phi80. J Mol Biol. 1966;19:254–265. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

