Crystal structure of the µ-opioid receptor bound to a morphinan antagonist
- PMID: 22437502
- PMCID: PMC3523197
- DOI: 10.1038/nature10954
Crystal structure of the µ-opioid receptor bound to a morphinan antagonist
Abstract
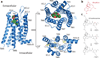
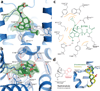
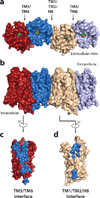
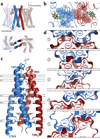
Opium is one of the world's oldest drugs, and its derivatives morphine and codeine are among the most used clinical drugs to relieve severe pain. These prototypical opioids produce analgesia as well as many undesirable side effects (sedation, apnoea and dependence) by binding to and activating the G-protein-coupled µ-opioid receptor (µ-OR) in the central nervous system. Here we describe the 2.8 Å crystal structure of the mouse µ-OR in complex with an irreversible morphinan antagonist. Compared to the buried binding pocket observed in most G-protein-coupled receptors published so far, the morphinan ligand binds deeply within a large solvent-exposed pocket. Of particular interest, the µ-OR crystallizes as a two-fold symmetrical dimer through a four-helix bundle motif formed by transmembrane segments 5 and 6. These high-resolution insights into opioid receptor structure will enable the application of structure-based approaches to develop better drugs for the management of pain and addiction.
Figures

Comment in
-
Structural biology: How opioid drugs bind to receptors.Nature. 2012 May 16;485(7398):314-7. doi: 10.1038/485314a. Nature. 2012. PMID: 22596150 Free PMC article.
References
-
- Katzung BG. Basic and clinical pharmacology. 10th edn. LANGE Mac Graw Hill Medical; 2007.
-
- Matthes HW, et al. Loss of morphine-induced analgesia, reward effect and withdrawal symptoms in mice lacking the mu-opioid-receptor gene. Nature. 1996;383:819–823. - PubMed
-
- Lord JA, Waterfield AA, Hughes J, Kosterlitz HW. Endogenous opioid peptides: multiple agonists and receptors. Nature. 1977;267:495–499. - PubMed
-
- Raffa RB, Martinez RP, Connelly CD. G-protein antisense oligodeoxyribonucleotides and mu-opioid supraspinal antinociception. Eur J Pharmacol. 1994;258:R5–R7. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

