Molecular characterization of feline infectious peritonitis virus strain DF-2 and studies of the role of ORF3abc in viral cell tropism
- PMID: 22438554
- PMCID: PMC3372193
- DOI: 10.1128/JVI.00189-12
Molecular characterization of feline infectious peritonitis virus strain DF-2 and studies of the role of ORF3abc in viral cell tropism
Abstract
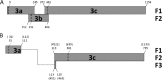
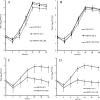
The full-length genome of the highly lethal feline infectious peritonitis virus (FIPV) strain DF-2 was sequenced and cloned into a bacterial artificial chromosome (BAC) to study the role of ORF3abc in the FIPV-feline enteric coronavirus (FECV) transition. The reverse genetic system allowed the replacement of the truncated ORF3abc of the original FIPV DF-2 genome with the intact ORF3abc of the canine coronavirus (CCoV) reference strain Elmo/02. The in vitro replication kinetics of these two viruses was studied in CrFK and FCWF-4 cell lines, as well as in feline peripheral blood monocytes. Both viruses showed similar replication kinetics in established cell lines. However, the strain with a full-length ORF3 showed markedly lower replication of more than 2 log(10) titers in feline peripheral blood monocytes. Our results suggest that the truncated ORF3abc plays an important role in the efficient macrophage/monocyte tropism of type II FIPV.
Figures


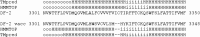


References
-
- Addie D, Jarrett O. 1992. A study of naturally occurring feline coronavirus infections in kittens. Vet. Rec. 130:133–137 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

