c-Myb regulates matrix metalloproteinases 1/9, and cathepsin D: implications for matrix-dependent breast cancer cell invasion and metastasis
- PMID: 22439866
- PMCID: PMC3325857
- DOI: 10.1186/1476-4598-11-15
c-Myb regulates matrix metalloproteinases 1/9, and cathepsin D: implications for matrix-dependent breast cancer cell invasion and metastasis
Abstract
Background: The c-Myb transcription factor is essential for the maintenance of stem-progenitor cells in bone marrow, colon epithelia, and neurogenic niches. c-Myb malfunction contributes to several types of malignancies including breast cancer. However, the function of c-Myb in the metastatic spread of breast tumors remains unexplored. In this study, we report a novel role of c-Myb in the control of specific proteases that regulate the matrix-dependent invasion of breast cancer cells.
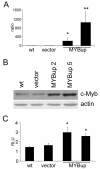
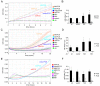
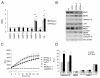
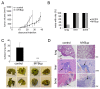
Results: Ectopically expressed c-Myb enhanced migration and ability of human MDA-MB-231 and mouse 4T1 mammary cancer cells to invade Matrigel but not the collagen I matrix in vitro. c-Myb strongly increased the expression/activity of cathepsin D and matrix metalloproteinase (MMP) 9 and significantly downregulated MMP1. The gene coding for cathepsin D was suggested as the c-Myb-responsive gene and downstream effector of the migration-promoting function of c-Myb. Finally, we demonstrated that c-Myb delayed the growth of mammary tumors in BALB/c mice and affected the metastatic potential of breast cancer cells in an organ-specific manner.
Conclusions: This study identified c-Myb as a matrix-dependent regulator of invasive behavior of breast cancer cells.
Figures

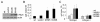
References
-
- Clappier E, Cuccuini W, Kalota A, Crinquette A, Cayuela JM, Dik WA, Langerak AW, Montpellier B, Nadel B, Walrafen P. et al.The C-MYB locus is involved in chromosomal translocation and genomic duplications in human T-cell acute leukemia (T-ALL), the translocation defining a new T-ALL subtype in very young children. Blood. 2007;110:1251–1261. doi: 10.1182/blood-2006-12-064683. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

