ARIEL and AMELIA: testing for an accumulation of rare variants using next-generation sequencing data
- PMID: 22441326
- PMCID: PMC3477640
- DOI: 10.1159/000336982
ARIEL and AMELIA: testing for an accumulation of rare variants using next-generation sequencing data
Abstract
Objectives: There is increasing evidence that rare variants play a role in some complex traits, but their analysis is not straightforward. Locus-based tests become necessary due to low power in rare variant single-point association analyses. In addition, variant quality scores are available for sequencing data, but are rarely taken into account. Here, we propose two locus-based methods that incorporate variant quality scores: a regression-based collapsing approach and an allele-matching method.
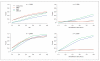
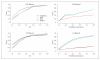
Methods: Using simulated sequencing data we compare 4 locus-based tests of trait association under different scenarios of data quality. We test two collapsing-based approaches and two allele-matching-based approaches, taking into account variant quality scores and ignoring variant quality scores. We implement the collapsing and allele-matching approaches accounting for variant quality in the freely available ARIEL and AMELIA software.
Results: The incorporation of variant quality scores in locus-based association tests has power advantages over weighting each variant equally. The allele-matching methods are robust to the presence of both protective and risk variants in a locus, while collapsing methods exhibit a dramatic loss of power in this scenario.
Conclusions: The incorporation of variant quality scores should be a standard protocol when performing locus-based association analysis on sequencing data. The ARIEL and AMELIA software implement collapsing and allele-matching locus association analysis methods, respectively, that allow the incorporation of variant quality scores.
Copyright © 2012 S. Karger AG, Basel.
Figures


References
-
- Cohen J, et al. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–872. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

