Intercellular variation in signaling through the TGF-β pathway and its relation to cell density and cell cycle phase
- PMID: 22442258
- PMCID: PMC3394940
- DOI: 10.1074/mcp.M111.013482
Intercellular variation in signaling through the TGF-β pathway and its relation to cell density and cell cycle phase
Abstract
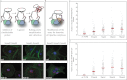
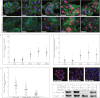
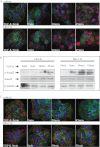
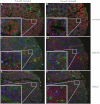
Fundamental open questions in signal transduction remain concerning the sequence and distribution of molecular signaling events among individual cells. In this work, we have characterized the intercellular variability of transforming growth factor β-induced Smad interactions, providing essential information about TGF-β signaling and its dependence on the density of cell populations and the cell cycle phase. By employing the recently developed in situ proximity ligation assay, we investigated the dynamics of interactions and modifications of Smad proteins and their partners under native and physiological conditions. We analyzed the kinetics of assembly of Smad complexes and the influence of cellular environment and relation to mitosis. We report rapid kinetics of formation of Smad complexes, including native Smad2-Smad3-Smad4 trimeric complexes, in a manner influenced by the rate of proteasomal degradation of these proteins, and we found a striking cell to cell variation of signaling complexes. The single-cell analysis of TGF-β signaling in genetically unmodified cells revealed previously unknown aspects of regulation of this pathway, and it provided a basis for analysis of these signaling events to diagnose pathological perturbations in patient samples and to evaluate their susceptibility to drug treatment.
Figures

References
-
- Massagué J., Blain S. W., Lo R. S. (2000) TGFβ signaling in growth control, cancer, and heritable disorders. Cell 103, 295–309 - PubMed
-
- Moustakas A., Heldin C. H. (2009) The regulation of TGFβ signal transduction. Development 136, 3699–3714 - PubMed
-
- Abdollah S., Macías-Silva M., Tsukazaki T., Hayashi H., Attisano L., Wrana J. L. (1997) TβRI phosphorylation of Smad2 on Ser465 and Ser467 is required for Smad2-Smad4 complex formation and signaling. J. Biol. Chem. 272, 27678–27685 - PubMed
-
- Akiyoshi S., Inoue H., Hanai J., Kusanagi K., Nemoto N., Miyazono K., Kawabata M. (1999) c-Ski acts as a transcriptional co-repressor in transforming growth factor-β signaling through interaction with smads. J. Biol. Chem. 274, 35269–35277 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

