Proteomic analysis of effluents from perfused human heart for transplantation: identification of potential biomarkers for ischemic heart damage
- PMID: 22443514
- PMCID: PMC3349588
- DOI: 10.1186/1477-5956-10-21
Proteomic analysis of effluents from perfused human heart for transplantation: identification of potential biomarkers for ischemic heart damage
Abstract
Background: Biomarkers released from the heart at early stage of ischemia are very important to diagnosis of ischemic heart disease and salvage myocytes from death. Known specific markers for blood tests including CK-MB, cardiac troponin T (cTnT) and cardiac troponin I (cTnI) are released after the onset of significant necrosis instead of early ischemia. Thus, they are not good biomarkers to diagnose myocardial injury before necrosis happens. Therefore, in this study, we performed proteomic analysis on effluents from perfused human hearts of donors at different ischemic time.
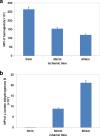
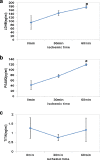
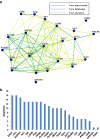
Results: After global ischemia for 0 min, 30 min and 60 min at 4°C, effluents from five perfused hearts were analyzed respectively, by High performance liquid chromatography-Chip-Mass spectrometry (HPLC-Chip-MS) system. Total 196 highly reliable proteins were identified. 107 proteins were identified at the beginning of ischemia, 174 and 175 proteins at ischemic 30 min and ischemic 60 min, respectively. With the exception of cardiac troponin I and T, all known biomarkers for myocardial ischemia were detected in our study. However, there were four glycolytic enzymes and two targets of matrix metalloproteinase released significantly from the heart when ischemic time was increasing. These proteins were L-lactate dehydrogenase B(LDHB), glyceraldehyde-3-phosphate dehydrogenase, glucose-6-phosphate isomerase (GPI), phosphoglycerate mutase 2 (PGAM2), gelsolin and isoform 8 of titin. PGAM2, LDHB and titin were measured with enzyme-linked immunosorbent assays kits. The mean concentrations of LDHB and PGAM2 in samples showed an increasing trend when ischemic time was extending. In addition, 33% identified proteins are involved in metabolism. Protein to protein interaction network analysis showed glycolytic enzymes, such as isoform alpha-enolase of alpha-enolase, isoform 1 of triosephosphate isomerase and glyceraldehyde-3-phosphate dehydrogenase, had more connections than other proteins in myocardial metabolism during ischemia.
Conclusion: It is the first time to use effluents of human perfused heart to study the proteins released during myocardial ischemia by HPLC-Chip-MS system. There might be many potential biomarkers for mild ischemic injury in myocardium, especially isoform 8 of titin and M-type of PGAM2 that are more specific in the cardiac tissue than in the others. Furthermore, glycolysis is one of the important conversions during early ischemia in myocardium. This finding may provide new insight into pathology and biology of myocardial ischemia, and potential diagnostic and therapeutic biomarkers.
Figures





References
-
- World Health Organization Department of Health Statistics and Informatics in the Information, Evidence and Research Cluster. The global burden of disease update. Geneva: WHO; 2004. ISBN 9241563710.
-
- Lewis GD, Wei R, Liu E, Yang E, Shi X, Martinovic M, Farrell L, Asnani A, Cyrille M, Ramanathan A. et al.Metabolite profiling of blood from individuals undergoing planned myocardial infarction reveals early markers of myocardial injury. J Clin Invest. 2008;118:3503–3512. doi: 10.1172/JCI35111. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

