The generation of chromosomal deletions to provide extensive coverage and subdivision of the Drosophila melanogaster genome
- PMID: 22445104
- PMCID: PMC3439972
- DOI: 10.1186/gb-2012-13-3-r21
The generation of chromosomal deletions to provide extensive coverage and subdivision of the Drosophila melanogaster genome
Abstract
Background: Chromosomal deletions are used extensively in Drosophila melanogaster genetics research. Deletion mapping is the primary method used for fine-scale gene localization. Effective and efficient deletion mapping requires both extensive genomic coverage and a high density of molecularly defined breakpoints across the genome.
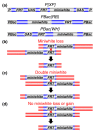
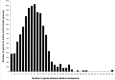
Results: A large-scale resource development project at the Bloomington Drosophila Stock Center has improved the choice of deletions beyond that provided by previous projects. FLP-mediated recombination between FRT-bearing transposon insertions was used to generate deletions, because it is efficient and provides single-nucleotide resolution in planning deletion screens. The 793 deletions generated pushed coverage of the euchromatic genome to 98.4%. Gaps in coverage contain haplolethal and haplosterile genes, but the sizes of these gaps were minimized by flanking these genes as closely as possible with deletions. In improving coverage, a complete inventory of haplolethal and haplosterile genes was generated and extensive information on other haploinsufficient genes was compiled. To aid mapping experiments, a subset of deletions was organized into a Deficiency Kit to provide maximal coverage efficiently. To improve the resolution of deletion mapping, screens were planned to distribute deletion breakpoints evenly across the genome. The median chromosomal interval between breakpoints now contains only nine genes and 377 intervals contain only single genes.
Conclusions: Drosophila melanogaster now has the most extensive genomic deletion coverage and breakpoint subdivision as well as the most comprehensive inventory of haploinsufficient genes of any multicellular organism. The improved selection of chromosomal deletion strains will be useful to nearly all Drosophila researchers.
Figures
References
-
- Parks AL, Cook KR, Belvin M, Dompe NA, Fawcett R, Huppert K, Tan LR, Winter CG, Bogart KP, Deal JE, Deal-Herr ME, Grant D, Marcinko M, Miyazaki WY, Robertson S, Shaw KJ, Tabios M, Vysotskaia V, Zhao L, Andrade RS, Edgar KA, Howie E, Killpack K, Milash B, Norton A, Thao D, Whittaker K, Winner MA, Friedman L, Margolis J. et al.Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat Genet. 2004;36:288–292. doi: 10.1038/ng1312. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

