An engineered eukaryotic protein glycosylation pathway in Escherichia coli
- PMID: 22446837
- PMCID: PMC3449280
- DOI: 10.1038/nchembio.921
An engineered eukaryotic protein glycosylation pathway in Escherichia coli
Abstract
We performed bottom-up engineering of a synthetic pathway in Escherichia coli for the production of eukaryotic trimannosyl chitobiose glycans and the transfer of these glycans to specific asparagine residues in target proteins. The glycan biosynthesis was enabled by four eukaryotic glycosyltransferases, including the yeast uridine diphosphate-N-acetylglucosamine transferases Alg13 and Alg14 and the mannosyltransferases Alg1 and Alg2. By including the bacterial oligosaccharyltransferase PglB from Campylobacter jejuni, we successfully transferred glycans to eukaryotic proteins.
Conflict of interest statement
Figures
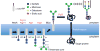
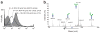

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

