Runx1 deletion or dominant inhibition reduces Cebpa transcription via conserved promoter and distal enhancer sites to favor monopoiesis over granulopoiesis
- PMID: 22451420
- PMCID: PMC3362359
- DOI: 10.1182/blood-2011-12-397091
Runx1 deletion or dominant inhibition reduces Cebpa transcription via conserved promoter and distal enhancer sites to favor monopoiesis over granulopoiesis
Abstract
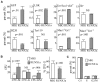
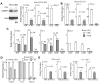
Deletion of Runx1 in adult mice produces a myeloproliferative phenotype. We now find that Runx1 gene deletion increases marrow monocyte while reducing granulocyte progenitors and that exogenous RUNX1 rescues granulopoiesis. Deletion of Runx1 reduces Cebpa mRNA in lineage-negative marrow cells and in granulocyte-monocyte progenitors or common myeloid progenitors. Pu.1 mRNA is also decreased, but to a lesser extent. We also transduced marrow with dominant-inhibitory RUNX1a. As with Runx1 gene deletion, RUNX1a expands lineage-Sca-1+c-kit+ and myeloid cells, increased monocyte CFUs relative to granulocyte CFUs, and reduced Cebpa mRNA. Runx1 binds a conserved site in the Cebpa promoter and binds 4 sites in a conserved 450-bp region located at +37 kb; mutation of the enhancer sites reduces activity 6-fold in 32Dcl3 myeloid cells. Endogenous Runx1 binds the promoter and putative +37 kb enhancer as assessed by ChIP, and RUNX1-ER rapidly induces Cebpa mRNA in these cells, even in cycloheximide, consistent with direct gene regulation. The +37 kb region contains strong H3K4me1 histone modification and p300-binding, as often seen with enhancers. Finally, exogenous C/EBPα increases granulocyte relative to monocyte progenitors in Runx1-deleted marrow cells. Diminished CEBPA transcription and consequent impairment of myeloid differentiation may contribute to leukemic transformation in acute myeloid leukemia cases associated with decreased RUNX1 activity.
Figures

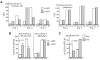


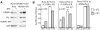
References
-
- Ichikawa M, Asai T, Saito T, et al. AML-1 is required for megakaryocytic maturation and lymphocytic differentiation, but not for maintenance of hematopoietic stem cells in adult hematopoiesis. Nat Med. 2004;10(3):299–304. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

