Klotho coreceptors inhibit signaling by paracrine fibroblast growth factor 8 subfamily ligands
- PMID: 22451487
- PMCID: PMC3347405
- DOI: 10.1128/MCB.06603-11
Klotho coreceptors inhibit signaling by paracrine fibroblast growth factor 8 subfamily ligands
Abstract
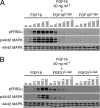
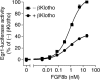
It has been recently established that Klotho coreceptors associate with fibroblast growth factor (FGF) receptor tyrosine kinases (FGFRs) to enable signaling by endocrine-acting FGFs. However, the molecular interactions leading to FGF-FGFR-Klotho ternary complex formation remain incompletely understood. Here, we show that in contrast to αKlotho, βKlotho binds its cognate endocrine FGF ligand (FGF19 or FGF21) and FGFR independently through two distinct binding sites. FGF19 and FGF21 use their respective C-terminal tails to bind to a common binding site on βKlotho. Importantly, we also show that Klotho coreceptors engage a conserved hydrophobic groove in the immunoglobulin-like domain III (D3) of the "c" splice isoform of FGFR. Intriguingly, this hydrophobic groove is also used by ligands of the paracrine-acting FGF8 subfamily for receptor binding. Based on this binding site overlap, we conclude that while Klotho coreceptors enhance binding affinity of FGFR for endocrine FGFs, they actively suppress binding of FGF8 subfamily ligands to FGFR.
Figures




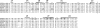

References
-
- ADHR Consortium 2000. Autosomal dominant hypophosphataemic rickets is associated with mutations in FGF23. Nat. Genet. 26:345–348 - PubMed
-
- Antoine M, et al. 2006. Fibroblast growth factor 16 and 18 are expressed in human cardiovascular tissues and induce on endothelial cells migration but not proliferation. Biochem. Biophys. Res. Commun. 246:224–233 - PubMed
-
- Asada M, et al. 2009. Glycosaminoglycan affinity of the complete fibroblast growth factor family. Biochim. Biophys. Acta 1790:40–48 - PubMed
-
- Badman MK, et al. 2007. Hepatic fibroblast growth factor 21 is regulated by PPARalpha and is a key mediator of hepatic lipid metabolism in ketotic states. Cell Metab. 5:426–437 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
