A dual role for the dREAM/MMB complex in the regulation of differentiation-specific E2F/RB target genes
- PMID: 22451490
- PMCID: PMC3372228
- DOI: 10.1128/MCB.06314-11
A dual role for the dREAM/MMB complex in the regulation of differentiation-specific E2F/RB target genes
Abstract
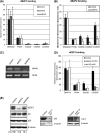
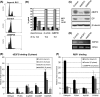
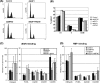
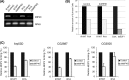
E2F and RB proteins regulate the expression of genes involved in cell cycle progression, apoptosis, differentiation, and development. Recent studies indicate that they function as part of an evolutionarily conserved multiprotein complex termed dREAM/DREAM/LINC. Here we characterize the role of the Drosophila complex, dREAM, in the regulation of differentiation-specific E2F target genes in actively proliferating cells. These genes are regulated differently from cell cycle-related E2F targets, they do not depend on E2F activation, and E2F/RB repression is maintained throughout the cell cycle. In proliferating cells, their repression is dependent on dREAM. We find that dREAM plays a dual role in their regulation. First, it is required for the stability of the repressive dE2F2/RBF complexes at their promoters during S phase. Second, we find that dREAM is indispensable for both transcriptional repression mechanisms employed at these genes.
Figures


Similar articles
-
Drosophila RB proteins repress differentiation-specific genes via two different mechanisms.Mol Cell Biol. 2010 May;30(10):2563-77. doi: 10.1128/MCB.01075-09. Epub 2010 Feb 22. Mol Cell Biol. 2010. PMID: 20176807 Free PMC article.
-
HDAC activity is dispensable for repression of cell-cycle genes by DREAM and E2F:RB complexes.Nat Commun. 2024 May 24;15(1):4450. doi: 10.1038/s41467-024-48724-0. Nat Commun. 2024. PMID: 38789411 Free PMC article.
-
p55, the Drosophila ortholog of RbAp46/RbAp48, is required for the repression of dE2F2/RBF-regulated genes.Mol Cell Biol. 2004 Oct;24(20):9124-36. doi: 10.1128/MCB.24.20.9124-9136.2004. Mol Cell Biol. 2004. PMID: 15456884 Free PMC article.
-
Cell cycle regulation: p53-p21-RB signaling.Cell Death Differ. 2022 May;29(5):946-960. doi: 10.1038/s41418-022-00988-z. Epub 2022 Mar 31. Cell Death Differ. 2022. PMID: 35361964 Free PMC article. Review.
-
Cell cycle transcription control: DREAM/MuvB and RB-E2F complexes.Crit Rev Biochem Mol Biol. 2017 Dec;52(6):638-662. doi: 10.1080/10409238.2017.1360836. Epub 2017 Aug 11. Crit Rev Biochem Mol Biol. 2017. PMID: 28799433 Review.
Cited by
-
The CHR site: definition and genome-wide identification of a cell cycle transcriptional element.Nucleic Acids Res. 2014;42(16):10331-50. doi: 10.1093/nar/gku696. Epub 2014 Aug 8. Nucleic Acids Res. 2014. PMID: 25106871 Free PMC article.
-
Novel functions for the transcription factor E2F4 in development and disease.Cell Cycle. 2016 Dec;15(23):3183-3190. doi: 10.1080/15384101.2016.1234551. Epub 2016 Oct 18. Cell Cycle. 2016. PMID: 27753528 Free PMC article. Review.
-
RBF binding to both canonical E2F targets and noncanonical targets depends on functional dE2F/dDP complexes.Mol Cell Biol. 2012 Nov;32(21):4375-87. doi: 10.1128/MCB.00536-12. Epub 2012 Aug 27. Mol Cell Biol. 2012. PMID: 22927638 Free PMC article.
-
A Cyclin A-Myb-MuvB-Aurora B network regulates the choice between mitotic cycles and polyploid endoreplication cycles.PLoS Genet. 2019 Jul 10;15(7):e1008253. doi: 10.1371/journal.pgen.1008253. eCollection 2019 Jul. PLoS Genet. 2019. PMID: 31291240 Free PMC article.
-
Loss of the mammalian DREAM complex deregulates chondrocyte proliferation.Mol Cell Biol. 2014 Jun;34(12):2221-34. doi: 10.1128/MCB.01523-13. Epub 2014 Apr 7. Mol Cell Biol. 2014. PMID: 24710275 Free PMC article.
References
-
- Beall EL, et al. 2002. Role for a Drosophila Myb-containing protein complex in site-specific DNA replication. Nature 420:833–837 - PubMed
-
- Cam H, Dynlacht BD. 2003. Emerging roles for E2F: beyond the G1/S transition and DNA replication. Cancer Cell 3:311–316 - PubMed
-
- Chan HM, Krstic-Demonacos M, Smith L, Demonacos C, La Thangue NB. 2001. Acetylation control of the retinoblastoma tumour-suppressor protein. Nat. Genet. 3:667–674 - PubMed
-
- Chen CR, Kang Y, Siegel PM, Massague J. 2002. E2F4/5 and p107 as Smad cofactors linking the TGFbeta receptor to c-myc repression. Cell 110:19–32 - PubMed
-
- Classon M, Harlow E. 2002. The retinoblastoma tumour suppressor in development and cancer. Nat. Rev. Cancer 2:910–917 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
