Cathepsins L and Z are critical in degrading polyglutamine-containing proteins within lysosomes
- PMID: 22451661
- PMCID: PMC3366842
- DOI: 10.1074/jbc.M112.352781
Cathepsins L and Z are critical in degrading polyglutamine-containing proteins within lysosomes
Abstract
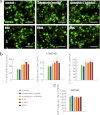
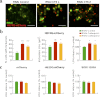
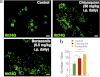
In neurodegenerative diseases caused by extended polyglutamine (polyQ) sequences in proteins, aggregation-prone polyQ proteins accumulate in intraneuronal inclusions. PolyQ proteins can be degraded by lysosomes or proteasomes. Proteasomes are unable to hydrolyze polyQ repeat sequences, and during breakdown of polyQ proteins, they release polyQ repeat fragments for degradation by other cellular enzymes. This study was undertaken to identify the responsible proteases. Lysosomal extracts (unlike cytosolic enzymes) were found to rapidly hydrolyze polyQ sequences in peptides, proteins, or insoluble aggregates. Using specific inhibitors against lysosomal proteases, enzyme-deficient extracts, and pure cathepsins, we identified cathepsins L and Z as the lysosomal cysteine proteases that digest polyQ proteins and peptides. RNAi for cathepsins L and Z in different cell lines and adult mouse muscles confirmed that they are critical in degrading polyQ proteins (expanded huntingtin exon 1) but not other types of aggregation-prone proteins (e.g. mutant SOD1). Therefore, the activities of these two lysosomal cysteine proteases are important in host defense against toxic accumulation of polyQ proteins.
Figures





References
-
- Zoghbi H. Y., Orr H. T. (2000) Glutamine repeats and neurodegeneration. Annu. Rev. Neurosci. 23, 217–247 - PubMed
-
- Rubinsztein D. C. (2006) The roles of intracellular protein-degradation pathways in neurodegeneration. Nature 443, 780–786 - PubMed
-
- Kitamura A., Kubota H. (2010) Amyloid oligomers. Dynamics and toxicity in the cytosol and nucleus. FEBS J. 277, 1369–1379 - PubMed
-
- Davies S. W., Turmaine M., Cozens B. A., DiFiglia M., Sharp A. H., Ross C. A., Scherzinger E., Wanker E. E., Mangiarini L., Bates G. P. (1997) Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell 90, 537–548 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

