Structure and mechanical properties of the ribosomal L1 stalk three-way junction
- PMID: 22451682
- PMCID: PMC3401443
- DOI: 10.1093/nar/gks258
Structure and mechanical properties of the ribosomal L1 stalk three-way junction
Abstract
The L1 stalk is a key mobile element of the large ribosomal subunit which interacts with tRNA during translocation. Here, we investigate the structure and mechanical properties of the rRNA H76/H75/H79 three-way junction at the base of the L1 stalk from four different prokaryotic organisms. We propose a coarse-grained elastic model and parameterize it using large-scale atomistic molecular dynamics simulations. Global properties of the junction are well described by a model in which the H76 helix is represented by a straight, isotropically flexible elastic rod, while the junction core is represented by an isotropically flexible spherical hinge. Both the core and the helix contribute substantially to the overall H76 bending fluctuations. The presence of wobble pairs in H76 does not induce any increased flexibility or anisotropy to the helix. The half-closed conformation of the L1 stalk seems to be accessible by thermal fluctuations of the junction itself, without any long-range allosteric effects. Bending fluctuations of H76 with a bulge introduced in it suggest a rationale for the precise position of the bulge in eukaryotes. Our elastic model can be generalized to other RNA junctions found in biological systems or in nanotechnology.
Figures



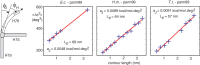
 is defined by Equation (6). The model predicts a linear relation between the contour length and the bending angle variation [Equation (5)]. The simulated data (blue crosses) satisfy the linear relation very well. The model stiffness parameters are inferred from the fitted linear functions (red lines). The intersection of the line with the y-axis determines the stiffness of the junction core aJ, the line slope determines the stiffness of the helix expressed by persistence length Lp [Equation (5) and (7)]. The stiffness parameters inferred from the data are shown. The complete list of the inferred parameters is in Table 1, analogous plots for the remaining systems are in
is defined by Equation (6). The model predicts a linear relation between the contour length and the bending angle variation [Equation (5)]. The simulated data (blue crosses) satisfy the linear relation very well. The model stiffness parameters are inferred from the fitted linear functions (red lines). The intersection of the line with the y-axis determines the stiffness of the junction core aJ, the line slope determines the stiffness of the helix expressed by persistence length Lp [Equation (5) and (7)]. The stiffness parameters inferred from the data are shown. The complete list of the inferred parameters is in Table 1, analogous plots for the remaining systems are in References
-
- Rodnina MV, Wintermeyer W, Green R. Ribosomes. Structure, Function, and Dynamics. Springer, Wien, New York. 2011
-
- Schmeing TM, Ramakrishnan V. What recent ribosome structures have revealed about the mechanism of translation. Nature. 2009;461:1234–1242. - PubMed

