An integrative probabilistic model for identification of structural variation in sequencing data
- PMID: 22452995
- PMCID: PMC3439973
- DOI: 10.1186/gb-2012-13-3-r22
An integrative probabilistic model for identification of structural variation in sequencing data
Abstract
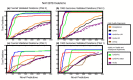
Paired-end sequencing is a common approach for identifying structural variation (SV) in genomes. Discrepancies between the observed and expected alignments indicate potential SVs. Most SV detection algorithms use only one of the possible signals and ignore reads with multiple alignments. This results in reduced sensitivity to detect SVs, especially in repetitive regions. We introduce GASVPro, an algorithm combining both paired read and read depth signals into a probabilistic model which can analyze multiple alignments of reads. GASVPro outperforms existing methods with a 50-90% improvement in specificity on deletions and a 50% improvement on inversions.
Figures

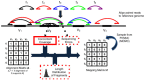
 from a test genome are sequenced and the resulting paired reads are aligned to the reference. A fragment may either have a unique mapping or be ambiguous with multiple alignments to the reference. Following clustering of alignments (with GASV), the set
from a test genome are sequenced and the resulting paired reads are aligned to the reference. A fragment may either have a unique mapping or be ambiguous with multiple alignments to the reference. Following clustering of alignments (with GASV), the set  of possible structural variants and the fragments whose alignments support these variants are recorded in the alignment matrix A. As each fragment originates from a single location in the test genome, a fragment supports at most one structural variant. Thus, the mapping matrix M records the 'true' mapping for each fragment. GASVPro scores mapping matrices according to a generative probabilistic model that incorporates concordant mappings. GASVPro utilizes an MCMC procedure to efficiently sample over the space of possible mapping matrices defined by the alignment matrix A. The underlying probabilistic model can be easily generalized to consider additional features indicative of a 'true' mapping, such as the empirical fragment length distribution or probability of sequencing errors.
of possible structural variants and the fragments whose alignments support these variants are recorded in the alignment matrix A. As each fragment originates from a single location in the test genome, a fragment supports at most one structural variant. Thus, the mapping matrix M records the 'true' mapping for each fragment. GASVPro scores mapping matrices according to a generative probabilistic model that incorporates concordant mappings. GASVPro utilizes an MCMC procedure to efficiently sample over the space of possible mapping matrices defined by the alignment matrix A. The underlying probabilistic model can be easily generalized to consider additional features indicative of a 'true' mapping, such as the empirical fragment length distribution or probability of sequencing errors.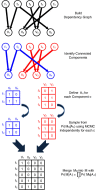


References
-
- Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y, Aerts J, Andrews TD, Barnes C, Campbell P, Fitzgerald T, Hu M, Ihm CH, Kristiansson K, Macarthur DG, Macdonald JR, Onyiah I, Pang AW, Robson S, Stirrups K, Valsesia A, Walter K, Wei J. Wellcome Trust Case Control Consortium. Tyler-Smith C, Carter NP, Lee C, Scherer SW, Hurles ME. Origins and functional impact of copy number variation in the human genome. Nature. 2009;464:704–712. - PMC - PubMed
-
- Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW, Harris CC, McLellan MD, Fulton RS, Fulton LL, Abbott RM, Hoog J, Dooling DJ, Koboldt DC, Schmidt H, Kalicki J, Zhang Q, Chen L, Lin L, Wendl MC, McMichael JF, Magrini VJ, Cook L, McGrath SD, Vickery TL, Appelbaum E, Deschryver K, Davies S, Guintoli T, Lin L. et al.Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989. - DOI - PMC - PubMed
-
- Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, Varela I, Lin ML, Ordóñez GR, Bignell GR, Ye K, Alipaz J, Bauer MJ, Beare D, Butler A, Carter RJ, Chen L, Cox AJ, Edkins S, Kokko-Gonzales PI, Gormley NA, Grocock RJ, Haudenschild CD, Hims MM, James T, Jia M, Kingsbury Z, Leroy C, Marshall J, Menzies A. et al.A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2009;463:191–196. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

