Epigenetic variability in cells of normal cytology is associated with the risk of future morphological transformation
- PMID: 22453031
- PMCID: PMC3446274
- DOI: 10.1186/gm323
Epigenetic variability in cells of normal cytology is associated with the risk of future morphological transformation
Abstract
Background: Recently, it has been proposed that epigenetic variation may contribute to the risk of complex genetic diseases like cancer. We aimed to demonstrate that epigenetic changes in normal cells, collected years in advance of the first signs of morphological transformation, can predict the risk of such transformation.
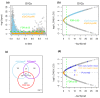
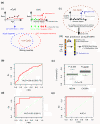
Methods: We analyzed DNA methylation (DNAm) profiles of over 27,000 CpGs in cytologically normal cells of the uterine cervix from 152 women in a prospective nested case-control study. We used statistics based on differential variability to identify CpGs associated with the risk of transformation and a novel statistical algorithm called EVORA (Epigenetic Variable Outliers for Risk prediction Analysis) to make predictions.
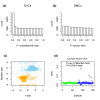
Results: We observed many CpGs that were differentially variable between women who developed a non-invasive cervical neoplasia within 3 years of sample collection and those that remained disease-free. These CpGs exhibited heterogeneous outlier methylation profiles and overlapped strongly with CpGs undergoing age-associated DNA methylation changes in normal tissue. Using EVORA, we demonstrate that the risk of cervical neoplasia can be predicted in blind test sets (AUC = 0.66 (0.58 to 0.75)), and that assessment of DNAm variability allows more reliable identification of risk-associated CpGs than statistics based on differences in mean methylation levels. In independent data, EVORA showed high sensitivity and specificity to detect pre-invasive neoplasia and cervical cancer (AUC = 0.93 (0.86 to 1) and AUC = 1, respectively).
Conclusions: We demonstrate that the risk of neoplastic transformation can be predicted from DNA methylation profiles in the morphologically normal cell of origin of an epithelial cancer. Having profiled only 0.1% of CpGs in the human genome, studies of wider coverage are likely to yield improved predictive and diagnostic models with the accuracy needed for clinical application.
Trial registration: The ARTISTIC trial is registered with the International Standard Randomised Controlled Trial Number ISRCTN25417821.
Figures



References
-
- Bibikova M, Fan JB. Genome-wide DNA methylation profiling. Wiley Interdiscip Rev Syst Biol Med. 2010;2:210–223. - PubMed
-
- Kitchener HC, Almonte M, Gilham C, Dowie R, Stoykova B, Sargent A, Roberts C, Desai M, Peto J. ARTISTIC: a randomised trial of human papillomavirus (HPV) testing in primary cervical screening. Health Technol Assess. 2009;13:1–150, iii-iv. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

