Folding without charges
- PMID: 22454493
- PMCID: PMC3326513
- DOI: 10.1073/pnas.1118640109
Folding without charges
Abstract
Surface charges of proteins have in several cases been found to function as "structural gatekeepers," which avoid unwanted interactions by negative design, for example, in the control of protein aggregation and binding. The question is then if side-chain charges, due to their desolvation penalties, play a corresponding role in protein folding by avoiding competing, misfolded traps? To find out, we removed all 32 side-chain charges from the 101-residue protein S6 from Thermus thermophilus. The results show that the charge-depleted S6 variant not only retains its native structure and cooperative folding transition, but folds also faster than the wild-type protein. In addition, charge removal unleashes pronounced aggregation on longer timescales. S6 provides thus an example where the bias toward native contacts of a naturally evolved protein sequence is independent of charges, and point at a fundamental difference in the codes for folding and intermolecular interaction: specificity in folding is governed primarily by hydrophobic packing and hydrogen bonding, whereas solubility and binding relies critically on the interplay of side-chain charges.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




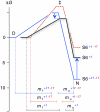
 ). The origin of this change could be the population of an alternative, parallel, folding route to the native state according to Scheme 2.
). The origin of this change could be the population of an alternative, parallel, folding route to the native state according to Scheme 2.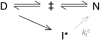
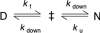
Similar articles
-
Gatekeepers in the ribosomal protein s6: thermodynamics, kinetics, and folding pathways revealed by a minimalist protein model.J Mol Biol. 2004 Jul 9;340(3):571-85. doi: 10.1016/j.jmb.2004.04.073. J Mol Biol. 2004. PMID: 15210355
-
Stabilization of the ribosomal protein S6 by trehalose is counterbalanced by the formation of a putative off-pathway species.J Mol Biol. 2005 Aug 12;351(2):402-16. doi: 10.1016/j.jmb.2005.05.056. J Mol Biol. 2005. PMID: 16002092
-
Simulation, experiment, and evolution: understanding nucleation in protein S6 folding.Proc Natl Acad Sci U S A. 2004 Jun 1;101(22):8354-9. doi: 10.1073/pnas.0401672101. Epub 2004 May 18. Proc Natl Acad Sci U S A. 2004. PMID: 15150413 Free PMC article.
-
Protein folding simulations: from coarse-grained model to all-atom model.IUBMB Life. 2009 Jun;61(6):627-43. doi: 10.1002/iub.223. IUBMB Life. 2009. PMID: 19472192 Review.
-
A Simple Principle for Understanding the Combined Cellular Protein Folding and Aggregation.Curr Protein Pept Sci. 2020;21(1):3-21. doi: 10.2174/1389203720666190725114550. Curr Protein Pept Sci. 2020. PMID: 31345145 Review.
Cited by
-
Atomic Resolution Structure of Monomorphic Aβ42 Amyloid Fibrils.J Am Chem Soc. 2016 Aug 3;138(30):9663-74. doi: 10.1021/jacs.6b05129. Epub 2016 Jul 14. J Am Chem Soc. 2016. PMID: 27355699 Free PMC article.
-
A Conceptual Framework for Integrating Cellular Protein Folding, Misfolding and Aggregation.Life (Basel). 2021 Jun 24;11(7):605. doi: 10.3390/life11070605. Life (Basel). 2021. PMID: 34202456 Free PMC article. Review.
-
Unified understanding of folding and binding mechanisms of globular and intrinsically disordered proteins.Biophys Rev. 2018 Apr;10(2):163-181. doi: 10.1007/s12551-017-0346-7. Epub 2018 Jan 6. Biophys Rev. 2018. PMID: 29307002 Free PMC article. Review.
-
An effective coarse-grained model for biological simulations: recent refinements and validations.Proteins. 2014 Jul;82(7):1168-85. doi: 10.1002/prot.24482. Proteins. 2014. PMID: 25050439 Free PMC article.
-
Visualization of protein folding funnels in lattice models.PLoS One. 2014 Jul 10;9(7):e100861. doi: 10.1371/journal.pone.0100861. eCollection 2014. PLoS One. 2014. PMID: 25010343 Free PMC article.
References
-
- Capaldi AP, Kleanthous C, Radford SE. Im7 folding mechanism: misfolding on a path to the native state. Nat Struct Biol. 2002;9:209–216. - PubMed
-
- Jin W, Kambara O, Sasakawa H, Tamura A, Takada S. De novo design of foldable proteins with smooth folding funnel: automated negative design and experimental verification. Structure. 2003;11:581–590. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

