Genetic switchboard for synthetic biology applications
- PMID: 22454498
- PMCID: PMC3326468
- DOI: 10.1073/pnas.1203808109
Genetic switchboard for synthetic biology applications
Abstract
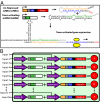
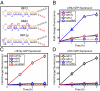
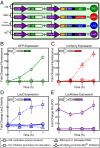
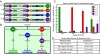
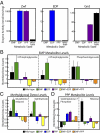
A key next step in synthetic biology is to combine simple circuits into higher-order systems. In this work, we expanded our synthetic riboregulation platform into a genetic switchboard that independently controls the expression of multiple genes in parallel. First, we designed and characterized riboregulator variants to complete the foundation of the genetic switchboard; then we constructed the switchboard sensor, a testing platform that reported on quorum-signaling molecules, DNA damage, iron starvation, and extracellular magnesium concentration in single cells. As a demonstration of the biotechnological potential of our synthetic device, we built a metabolism switchboard that regulated four metabolic genes, pgi, zwf, edd, and gnd, to control carbon flow through three Escherichia coli glucose-utilization pathways: the Embden-Meyerhof, Entner-Doudoroff, and pentose phosphate pathways. We provide direct evidence for switchboard-mediated shunting of metabolic flux by measuring mRNA levels of the riboregulated genes, shifts in the activities of the relevant enzymes and pathways, and targeted changes to the E. coli metabolome. The design, testing, and implementation of the genetic switchboard illustrate the successful construction of a higher-order system that can be used for a broad range of practical applications in synthetic biology and biotechnology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Ellis T, Adie T, Baldwin GS. DNA assembly for synthetic biology: From parts to pathways and beyond. Integr Biol (Camb) 2011;3:109–118. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

